Abstract
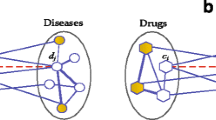
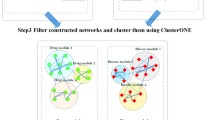
Computational techniques for foreseeing drug-disease associations by means of incorporating gene expression as well as biological network give high intuitions to the composite associations amongst targets, drugs, disease genes in addition to the diseases at a system level. Hepatocellular Carcinoma (HCC) is a malevolent tumor containing a greater rate of sickness as well as mortality. In the present work, an Integrative framework is presented with the aim of resolving this problem, for identifying new Drugs for HCC dependent upon Multi-Source Random Walk (PD-MRW), in which score the complete drugs by means of building the drug-drug similarity network. On the other hand, the collection of clinical phenotypes as well as drug side effects in combination with patient-specific genetic info. As a result, the formation of disease-drug networks that denotes the prescriptions, which are allotted to treat those diseases that are not concentrated by means of PD-MRW model. With the aim of overcoming this issue, this research offers an integrative framework for foreseeing new drugs as well as diseases for HCC dependent upon Multi-Source Simulated Annealing based Random Walk (PDD-MSSARW). Primarily, build a Gene-Gene Weighted Interaction Network (GWIN), dependent upon the gene expression as well as protein interaction network. After that, construct a drug-drug similarity network, dependent upon multi-source random walk in GWIN, disease-drug similarity network with the help of Similarity Weighted Bipartite Graph Network (SWBGN) that is build up in which the nodes are drugs as well as association among one node to another node that explains the disease diagnoses. Lastly, dependent upon the known drugs for HCC, score the entire drugs in the similarity networks. The sturdiness of the likelihoods, their overlap with those stated in Comparative Toxicogenomics Database (CTD) as well as kinds of literature, and their enhanced KEGG pathway illustrate PDD-MSSARW method be capable of efficiently find out novel drug signs.





Similar content being viewed by others
References
DiMasi, J., Hansen, R., and Grabowski, H., The price of innovation: New estimates of drug development costs. J. Health Econ. 22(2):151–185, 2003.
Chong, C., and Sullivan, D., New uses for old drugs. Nature 448(7154):645–646, 2007.
Pujol, A., Mosca, R., Farrés, J., and Aloy, P., Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. 31(3):115–123, 2010.
Jones, D., Pathways to cancer therapy. Nat. Rev. Drug Discov. 7(11):875–876, 2008.
Li, J., Zhu, X., and Chen, J. Y., Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS Comput. Biol. 5:e1000450, 2009.
Hurle, M. R., Yang, L., Xie, Q., Rajpal, D. K., Sanseau, P., and Agarwal, P., Computational drug repositioning: From data to therapeutics. Clin. Pharmacol. Ther. 93(4):335–341, 2013.
Duran-Frigola, M., and Aloy, P., Recycling side-effects into clinical markers for drug repositioning. Genome Med. 4:3, 2012.
Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P, "A Side Effect Re-source to capture phenotypic effects of drugs," Mol. Syst. Biol.. 6: 343, 2010.
Ye, H., Liu, Q., and Wei, J., Construction of drug network based on side effects and its application for drug repositioning. PLoS One 9(2):e87864, 2014.
Wang, W., Yang, S., Zhang, X., and Li, J., Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 30(20):2923–2930, 2014.
Gottlieb, A., Stein, G. Y., Ruppin, E., and Sharan, R., PREDICT a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 7(1):1–15, 2011.
Cheng, D., Knox, C., Young, N., Stothard, P., Damaraju, S., and Wishart, D., PolySearch: A web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs, and metabolites. Nucleic Acids Res. 36:W399–W405, 2008.
Ozgür, A., Vu, T., Erkan, G., and Radev, D., Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics 24(13):277–285, 2008.
Chen, X., Liu, M. X., and Yan, G. Y., Drug-target interaction prediction by random walk on the heterogeneous network. Mol. BioSyst. 8:1970–1978, 2012.
Liu, H., Song, Y., Guan, J., Luo, L., and Zhuang, Z., Inferring new indications for approved drugs via a random walk on drug-disease heterogenous networks. BMC Bioinform. 17(17):539, 2016.
Ba-Alawi, W., Soufan, O., Essack, M. et al., DASPfind: New efficient method to predict drug-target interactions. J. Cheminform. 8(1):15, 2016.
Alaimo, S., Pulvirenti, A., Giugno, R., and Ferro, A., Drug–target interaction prediction through domain-tuned network-based inference. Bioinformatics 29(16):2004–2008, 2013.
Hwang, T., and Kuang, R., "A Heterogeneous Label Propagation algorithm for disease gene discovery, “ In SIAM international conference on data mining, pp. 583–594, 2001.
Liu, Y., Wu, M., Miao, C. et al., Neighborhood regularized logistic matrix factorization for drug-target interaction prediction. PLoS. Comput. Biol. 12(2):e1004760, 2016.
Hao, M., Bryant, S. H., and Wang, Y., Predicting drug-target interactions by dual-network integrated logistic matrix factorization. Sci. Rep. 7:40376, 2017.
Wang, L., You, Z.-H., Chen, X. et al., RFDT: A rotation forest-based predictor for predicting drug-target interactions using drug structure and protein sequence information. Curr. Protein PeptSci. 18(999):1, 2016.
Ezzat, A., Wu, M., Li, X.-L. et al., Drug-target interaction prediction via class imbalance-aware ensemble learning. BMC Bioinform. 17(19):267–276, 2016.
Nascimento, A. C. A., RBC, P., and Costa, I. G., A multiple kernel learning algorithm for drug-target interaction prediction. BMC Bioinform. 17(1):46, 2016.
Yuan, Q., Gao, J., Wu, D. et al., DrugE-rank: Improving drug–target interaction prediction of new candidate drugs or targets by ensemble learning to rank. Bioinformatics 32(12):i18–i27, 2016.
Gawehn, E., Hiss, J. A., and Schneider, G., Deep learning in drug discovery. Mol. Inform. 35(1):3–14, 2016.
Ekins, S., The next era: Deep learning in pharmaceutical research. Pharm. Res. 33(11):2594–2603, 2016.
Abdel-Basset, M., El-Shahat, D., and Mirjalili, S., A hybrid whale optimization algorithm based on local search strategy for the permutation flow shop scheduling problem. Futur. Gener. Comput. Syst. 85:129–145, 2018.
Abdel-Basset, M., Abdel-Fatah, M.G.L., and Mirjalili, S., An improved nature inspired meta-heuristic algorithm for 1-D bin packing problems. Pers. Ubiquit. Comput., 1–16, 2018.
Abdel-Basset, M., Gamal, M.G.A., and Smarandache, F., A hybrid approach of neutrosophic sets and DEMATEL method for developing supplier selection criteria. Des. Autom. Embed. Syst., 1–22, 2018.
Abdel-Basset, M., Mohamed, M.G.M., and Smarandache, F. A novel method for solving the fully neutrosophic linear programming problems. Neural Comput. & Applic., 1–11.
Abdel-Basset, M., Fakhry, M.G.A.E., and El-Henawy, I., 2-Levels of clustering strategy to detect and locate copy-move forgery in digital images. Multimed. Tools Appl., 1–19, 2018.
Abdel-Basset, M., and Mohamed, M., Internet of things (IoT) and its impact on supply chain: A framework for building smart, secure and efficient systems. Futur. Gener. Comput. Syst., 2018.
Abdel-Basset, M., Manogaran, G., Mohamed, M., and Rushdy, E. Internet of things in smart education environment: a Supportive framework in the decision-making process. concurrency and Computation: Practice and Experience, e4515.
Abdel-Basset, M., Rashad, M.G.H., and Zaied, A.N.H., A comprehensive review of the quadratic assignment problem: Variants, hybrids, and applications. J. Ambient. Intell. Humaniz. Comput., 1–24, 2018.
Abdel-Basset, M., Gunasekaran, M., Mohamed, M., and Chilamkurti, N., Three-way decisions based on neutrosophic sets and AHP-QFD framework for supplier selection problem. Futur. Gener. Comput. Syst., 2018.
Davis, A. P., Grondin, C. J., Lennon-Hopkins, K., Saraceni-Richards, C., Sciaky, D., King, B. L., Wiegers, T. C., and Mattingly, C. J., The comparative Toxicogenomics Database's 10th year anniversary: Update 2015. Nucleic Acids Res. 43(D1):D914–D920, 2014.
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., Marshall, K. A., Phillippy, K. H., Sherman, P. M., Holko, M., and Yefanov, A., NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 41(D1):D991–D995, 2012.
Keshava Prasad, T. S., Goel, R., Kandasamy, K., Keerthikumar, S., Kumar, S., Mathivanan, S., Telikicherla, D., Raju, R., Shafreen, B., Venugopal, A., and Balakrishnan, L., Human protein reference database—2009 update. Nucleic Acids Res. 37(suppl_1):D767–D772, 2008.
Gilbert, D., Biomolecular interaction network database. Brief. Bioinform. 6(2):194–198, 2005.
Alfarano, C., Andrade, C. E., Anthony, K., Bahroos, N., Bajec, M., Bantoft, K., Betel, D., Bobechko, B., Boutilier, K., Burgess, E., and Buzadzija, K., The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res. 33(suppl_1):D418–D424, 2005.
Hermjakob, H., Montecchi-Palazzi, L., Lewington, C., Mudali, S., Kerrien, S., Orchard, S., Vingron, M., Roechert, B., Roepstorff, P., Valencia, A., and Margalit, H., IntAct: An open source molecular interaction database. Nucleic Acids Res. 32(suppl_1):D452–D455, 2004.
Kerrien, S., Aranda, B., Breuza, L., Bridge, A., Broackes-Carter, F., Chen, C., Duesbury, M., Dumousseau, M., Feuermann, M., Hinz, U., and Jandrasits, C., The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40(D1):D841–D846, 2011.
Ceol, A., ChatrAryamontri, A., Licata, L., Peluso, D., Briganti, L., Perfetto, L., Castagnoli, L., and Cesareni, G., MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 38(suppl_1):D532–D539, 2009.
Licata, L., Briganti, L., Peluso, D., Perfetto, L., Iannuccelli, M., Galeota, E., Sacco, F., Palma, A., Nardozza, A. P., Santonico, E., and Castagnoli, L., MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 40(D1):D857–D861, 2011.
Brown, K. R., and Jurisica, I., Online predicted human interaction database. Bioinformatics 21(9):2076–2082, 2005.
Meyer, M.J., Beltrán, J.F., Liang, S., Fragoza, R., Rumack, A., Liang, J., Wei, X. and Yu, H., 2018. Interactome INSIDER: a structural interactome browser for genomic studies. Nat. Methods, vol. 15, no.2 , pp. 107–120, 2018.
Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., Chang, Z., and Woolsey, J., DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34(suppl_1):D668–D672, 2006.
Wishart, D. S., Knox, C., Guo, A. C., Cheng, D., Shrivastava, S., Tzur, D., Gautam, B., and Hassanali, M., DrugBank: A knowledgebase for drugs, drug actions, and drug targets. Nucleic Acids Res. 36(suppl_1):D901–D906, 2007.
Knox, C., Law, V., Jewison, T., Liu, P., Ly, S., Frolkis, A., Pon, A., Banco, K., Mak, C., Neveu, V., and Djoumbou, Y., DrugBank 3.0: A comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 39(suppl_1):D1035–D1041, 2010.
Law, V., Knox, C., Djoumbou, Y., Jewison, T., Guo, A. C., Liu, Y., Maciejewski, A., Arndt, D., Wilson, M., Neveu, V., and Tang, A., DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 42(D1):D1091–D1097, 2013.
Huang, D. W., Sherman, B. T., and Lempicki, R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57, 2008.
Huang, D. W., Sherman, B. T., and Lempicki, R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13, 2009.
Frieze, A. M., Cooper, C., Siam, J., and Radzik, T., Multiple random walks in random regular graphs. Discret. Math. 23:1738–1761, 2009.
Aarts, E., Korst, J., and Michiels, W., Simulated annealing. Search methodologies. Boston, MA: Springer, 2005, 187–210.
Scarpa, G., Gaetano, R., Haindl, M., and Zerubia, J., Hierarchical multiple Markov chain model for unsupervised texture segmentation. IEEE Trans. Image Process. 18(8):1830–1843, 2009.
Menche, J., Sharma, A., Kitsak, M., Ghiassian, S. D., Vidal, M., Loscalzo, J., and Barabási, A. L., Uncovering disease-disease relationships through the incomplete interactome. Science 347(6224):1257601, 2015.
Li, Z., Wu, X.M. and Chang, S.F., Segmentation using superpixels: A bipartite graph partitioning approach. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 789–796, 2012.
Zhou, T., Ren, J., Medo, M., and Zhang, Y. C., Bipartite network projection and personal recommendation. Phys. Rev. E 76(4):046115, 2007.
Kathiresan, V. and Sumathi, P., An efficient clustering algorithm based on Z-score ranking method. International Conference on Computer Communication and Informatics (ICCCI), pp. 1–4, 2012.
Powers, D.M., 2011. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness, and correlation. J. Mach. Learn. Technol. Volume 2, Issue 1, 2011, pp-37-63.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Jafar Ali Ibrahim has no conflict of interest. Thangamani has no conflict of interest.
Ethical Approval
This Article Does not contain any studies with Human Participants or animals performed by any of the authors.
Informed Consent
As we are not used Human Participation no, such informed consent.
Additional information
This article is part of the Topical Collection on Patient Facing Systems
Rights and permissions
About this article
Cite this article
Ibrahim, S.J.A., Thangamani, M. Prediction of Novel Drugs and Diseases for Hepatocellular Carcinoma Based on Multi-Source Simulated Annealing Based Random Walk. J Med Syst 42, 188 (2018). https://doi.org/10.1007/s10916-018-1038-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-1038-y