Abstract
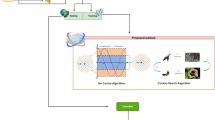
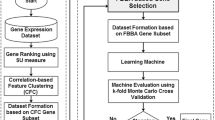
Microarray technology is utilized by the biologists, in order to compute the expression levels of thousands of genes. Cervical cancer classification utilizing gene expression data depends upon conventional supervised learning methods, wherein only labeled data could be used for learning. The previous methodologies had problem with appropriate feature selection as well as accurateness of classification outcomes. So, the entire performance of the cancer classification is decreased meaningfully. With the aim of overcoming the aforesaid problems, Enhanced Bat Optimization Algorithm with Hilbert-Schmidt Independence Criterion (EBO-HSIC) and Support Vector Machine (SVM) algorithm is presented in this research for identifying the specific genes from the gene expression dataset that belongs to cancer microarray. This proposed system contains phases of instance normalization, module detection, gene selection and classification. By Fuzzy C Means (FCM) algorithm, the normalization is performed for eliminating the inappropriate features from the gene dataset. Meanwhile, for effective feature selection, the EBO algorithm is used for producing more appropriate features via improved objective function values. For determining a subset of the most informative genes utilizing a rapid as well as scalable bat algorithm, this proposed method focuses on measuring the dependence amid Differentially Expressed Genes (DEGs) as well as the gene significance. The algorithm is dependent upon the HSIC and was partially enthused by EBO. With the help of SVM classifier, these gene features are categorized very precisely. Experimentation outcomes demonstrate that the presented EBO with SVM algorithm confirms a clear-cut classification performance for the given gene expression datasets. Hence the result provides higher performance by launching EBO with SVM algorithm to obtain greater accuracy, recall, precision, f-measure and less time complexity more willingly than the previous techniques.




Similar content being viewed by others
References
Denny, L., Cervical cancer: Prevention and treatment. Discov Med. 14:125–131, 2012.
Satija, A., Cervical cancer in India. South Asia Centre for chronic disease.[accessed February16, 2014], 2014. Available from: http://sancd.Org/uploads/ pdf/cervical_cancer.Pdf, 2.
Arbyn, M., Castellsague, X., DeSanjose, S. et al., Worldwide burden of cervical cancer. Ann. Oncol. 22:2675–2686, 2011.
Yeole, B. B., Kumar, A. V., Kurkureet, A., and Sunny, L., Population-based survival from cancers of breast, cervix and ovary in women in Mumbai. Asian Pac. J Cancer Prev. 5:308–315, 2004.
Bruni, L., Barrionuevo-Rosas, L., Albero, G., Serrano, B., Mena, M. and Gómez, D., ICO information Centre on HPV and Cancer. Human papillomavirus and related diseases in Ghana. Summary Report, HI Centre, Editor, 2015.
Gadducci, A., Barsotti, C., Cosio, S., Domenici, L., and Riccardo, A. G., Smoking habit, immune suppression, oral contraceptive use, and hormone replacement therapy use and cervical carcinogenesis: A review of the literature. Gynecol. Endocrinol. 27(8):597–604, 2011.
Stuart, C., and Ash, M., Gynaecology by ten teachers (18 ed.). London, U.K: Hodder education, 2006.
Croce, C. M., Oncogenes and cancer. N. Engl. J. Med. 358(5):502–511, 2008.
Wang, S. S., Gonzalez, P., Yu, K., Porras, C., Li, Q., Safaeian, M., Rodriguez, A. C., Sherman, M. E., Bratti, C., Schiffman, M., and Wacholder, S., Common genetic variants and risk for HPV persistence and progression to cervical cancer. PloS one 5(1):e8667, 2010.
Huang, D. S., and Yu, H. J., Normalized feature vectors: A novel alignment-free sequence comparison method based on the numbers of adjacent amino acids. IEEE/ACM Trans. Comput. Biol. Bioinformat. 10(2):457–467, 2013.
Wang, S. L., Zhu, Y., Jia, W., and Huang, D. S., Robust classification method of tumor subtype by using correlation filters. IEEE/ACM Trans. Comput. Biol. Bioinformat. 9(2):580–591, 2012.
Bergmann, S. et al., Similarities and differences in genome-wide expression data of six organisms. PLoSBiol 2:E9, 2004.
Hudson, N. J., Reverter, A., and Dalrymple, B. P., A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoSComput. Biol. 5(5):e1000382, 2009.
Maji, P., F-information measures for efficient selection of discriminative genes from microarray data. IEEE Trans. Biomed. Eng. 56(4):1063–1069, 2009.
Guyon, I., and Elisseeff, A., An introduction to variable and feature selection. J. Mach. Learn. Res. 3:1157–1182, 2003.
Peng, H., Long, F., and Ding, C., Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 27(8):1226–1238, 2005.
Cheng, Q., Zhou, H., and Cheng, J., The fisher-Markov selector: Fast selecting maximally separable feature subset for multiclass classification with applications to high-dimensional data. IEEE Trans. Pattern Anal. Mach. Intell. 33(6):1217–1233, 2011.
Lee, K. S., and Geem, Z. W., A new meta-heuristic algorithm for continuous engineering optimization: Harmony search theory and practice. Comput. Methods Appl .Mech. Eng. 194(36–38):3902–3933, 2005.
Yang, X.S., A new metaheuristic bat-inspired algorithm. Nature inspired cooperative strategies for optimization (NICSO 2010) (pp. 65–74). Springer, Berlin, Heidelberg, 2010.
Tang, E.K., Suganthan, P.N. and Yao, X., Feature selection for microarray data using least squares SVM and particle swarm optimization. IEEE Symp. Comput. Intell. Bioinform. Comput. Biol. 2005 (CIBCB'05), 1–8, 2005.
Gretton, A., Bousquet, O., Smola, A. and Schölkopf, B., Measuring statistical dependence with Hilbert-Schmidt norms. In International conference on algorithmic learning theory (pp. 63–77). Springer, Berlin, Heidelberg, 2005.
Hernandez, J. C., Duval, B., and Hao, J.-K., SVM-based local search for gene selection and classification of microarray data. Bioinform. Res. Dev. Springer, Berlin, Heidelberg. 499–508, 2008.
Chen, X., Jiang, J., Shen, H., and Hu, Z., Genetic susceptibility of cervical cancer. J. Biomed. Res. 25(3):155–164, 2011.
Thomas, A., Mahantshetty, U., Kannan, S., Deodhar, K., Shrivastava, S. K., Kumar-Sinha, C., and Mulherkar, R., Expression profiling of cervical cancers in Indian women at different stages to identify gene signatures during progression of the disease. Canc. Med 2(6):836–848, 2013.
Ongenaert, M., Wisman, G. B. A., Volders, H. H., Koning, A. J., van der Zee, A. G., Van Criekinge, W., and Schuuring, E., Discovery of DNA methylation markers in cervical cancer using relaxation ranking. BMC Med. Genom. 1(1):57, 2008.
Viswanathan, V. and Vineetha, S., Early detection of cervical cancer using microarray analysis and gene regulatory rules. International Conference on Emerging Technological Trends (ICETT), pp. 1–6, 2016.
Lee, H. S., Yun, J. H., Jung, J., Yang, Y., Kim, B. J., Lee, S. J., Yoon, J. H., Moon, Y., Kim, J. M., and Kwon, Y. I., Identification of differentially-expressed genes by DNA methylation in cervical cancer. Oncol. Lett. 9(4):1691–1698, 2015.
Mine, K. L., Shulzhenko, N., Yambartsev, A., Rochman, M., Sanson, G. F., Lando, M., Varma, S., Skinner, J., Volfovsky, N., Deng, T., and Brenna, S. M., Gene network reconstruction reveals cell cycle and antiviral genes as major drivers of cervical cancer. Nat. Commun. 4(1806):1–11, 2013.
Langfelder, P., and Horvath, S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9(1):1–13, 2008.
DiLeo, M. V., Strahan, G. D., den Bakker, M., and Hoekenga, O. A., Weighted correlation network analysis (WGCNA) applied to the tomato fruit metabolome. PLoS One 6(10):e26683, 2011.
Chuang, K. S., Tzeng, H. L., Chen, S., Wu, J., and Chen, T. J., Fuzzy c-means clustering with spatial information for image segmentation. Comput. Med. Imag. Graph. 30(1):9–15, 2006.
Zhang, S., Wang, R. S., and Zhang, X. S., Identification of overlapping community structure in complex networks using fuzzy c-means clustering. Phys. A: Stat. Mech. Appl. 374(1):483–490, 2007.
Van der Laan, M., Pollard, K., and Bryan, J., A new partitioning around medoids algorithm. J. Stat. Comput. Simul 73(8):575–584, 2003.
Langfelder, P., Zhang, B., and Horvath, S., Defining clusters from a hierarchical cluster tree: The dynamic tree cut package for R. Bioinformatics 24(5):719–720, 2007.
Rai, P., and Singh, S., A survey of clustering techniques. Int. J. Comput. Appl. 7(12):1–5, 2010.
Bhat, A., K-medoids clustering using partitioning around medoids for performing face recognition. Int. J. Soft Comput. Math. Contrl. 3(3):1–12, 2014.
Song, J. B., Borgwardt, K. M., Gretton, A., and Smola, A. J., Gene selection via the BAHSIC family of algorithms. Bioinf. 23:i490–i498, 2007.
Yang, X. S., and Hossein Gandomi, A., Bat algorithm: A novel approach for global engineering optimization. Eng. Comput. 29(5):464–483, 2012.
Gandomi, A. H., Yang, X. S., Alavi, A. H., and Talatahari, S., Bat algorithm for constrained optimization tasks. Neural Comput. Appl. 22(6):1239–1255, 2013.
Yang, X. S., Bat algorithm for multi-objective optimisation. Int. J. Bio-Inspired Comput. 3(5):267–274, 2011.
Spitzer, F., Principles of random walk (Vol. 34). Springer Science & Business Media, 2013.
Wang, L. Ed., 2005. Support vector machines: Theory and applications (Vol. 177). Springer Science & Business Media, 2005.
Fung, G. M., and Mangasarian, O. L., Multicategory proximal support vector machine classifiers. Mach. Learn. 59(1–2):77–97, 2005.
Min, J. H., and Lee, Y. C., Bankruptcy prediction using support vector machine with optimal choice of kernel function parameters. Expert Syst. Appl. 28(4):603–614, 2005.
Widodo, A., and Yang, B. S., Support vector machine in machine condition monitoring and fault diagnosis. Mech. Syst. Sign. Process. 21(6):2560–2574, 2007.
Sokolova, M., and Lapalme, G., A systematic analysis of performance measures for classification tasks. Inform. Process. Manag. 45(4):427–437, 2009.
García, S., Fernández, A., Luengo, J., and Herrera, F., A study of statistical techniques and performance measures for genetics-based machine learning: Accuracy and interpretability. Soft Comput. 13(10):959–977, 2009.
Pepe, M. S., Feng, Z., Janes, H., Bossuyt, P. M., and Potter, J. D., Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J. Natl. Cancer Instit. 100(20):1432–1438, 2008.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Transactional Processing Systems
Rights and permissions
About this article
Cite this article
Geeitha, S., Thangamani, M. Incorporating EBO-HSIC with SVM for Gene Selection Associated with Cervical Cancer Classification. J Med Syst 42, 225 (2018). https://doi.org/10.1007/s10916-018-1092-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-1092-5