Abstract
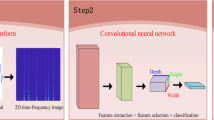
This study uses convolutional neural networks (CNNs) and cardiotocography data for the real-time classification of fetal status in the mobile application of a pregnant woman and the computer server of a data expert at the same time (The sensor is connected with the smartphone, which is linked with the web server for the woman and the computer server for the expert). Data came from 5249 (or 4833) cardiotocography traces in Anam Hospital for the mobile application (or the computer server). 150 data cases of 5-minute duration were extracted from each trace with 141,001 final cases for the mobile application and for the computer server alike. The dependent variable was fetal status with two categories (Normal, Abnormal) for the mobile application and three categories (Normal, Middle, Abnormal) for the computer server. The fetal heart rate served as a predictor for the mobile application and the computer server, while uterus contraction for the computer server only. The 1-dimension (or 2-dimension) Resnet CNN was trained for the mobile application (or the computer server) during 800 epochs. The sensitivity, specificity and their harmonic mean of the 1-dimension CNN for the mobile application were 94.9%, 91.2% and 93.0%, respectively. The corresponding statistics of the 2-dimension CNN for the computer server were 98.0%, 99.5% and 98.7%. The average inference time per 1000 images was 6.51 micro-seconds. Deep learning provides an efficient model for the real-time classification of fetal status in the mobile application and the computer server at the same time.





Similar content being viewed by others
Code and Data Availability
The code and data presented in this study are not publicly available. But the code and data are available from the corresponding author upon reasonable request.
References
U.S. Center for Disease Control. Pregnancy mortality surveillance system. 2022. Available online: https://www.cdc.gov/reproductivehealth/maternal-mortality/pregnancy-mortality-surveillance-system.htm (accessed on 1, June 2022).
Cleveland Clinic. Health library: high-risk pregnancy. 2022. Available online: https://my.clevelandclinic.org/health/diseases/22190-high-risk-pregnancy (accessed on 1 June 2022).
Mayo Clinic. Healthy lifestyle: high-risk pregnancy. 2022. Available online: https://www.mayoclinic.org/healthy-lifestyle/pregnancy-week-by-week/in-depth/high-risk-pregnancy/art-20047012 (accessed on 1 June 2022).
Mathews TJ, Driscoll AK. Trends in infant mortality in the United States, 2005–2014. NCHS data brief, no 279. Hyattsville, MD: National Center for Health Statistics. 2017.
American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics. Antepartum Fetal Surveillance: ACOG Practice Bulletin, Number 229. Obstet Gynecol. 2021 Jun 1;137(6):e116-e127.
Ayres-de-Campos D. Electronic fetal monitoring or cardiotocography, 50 years later: what’s in a name? Am J Obstet Gynecol. 2018 Jun;218(6):545–546.
ACOG Practice Bulletin No. 106: Intrapartum fetal heart rate monitoring: nomenclature, interpretation, and general management principles. Obstet Gynecol. 2009 Jul;114(1):192–202.
Macones GA, Hankins GD, Spong CY, Hauth J, Moore T. The 2008 National Institute of Child Health and Human Development workshop report on electronic fetal monitoring: update on definitions, interpretation, and research guidelines. Obstet Gynecol. 2008 Sep;112(3):661–6.
Ayres-de-Campos D, Spong CY, Chandraharan E; FIGO Intrapartum Fetal Monitoring Expert Consensus Panel. FIGO consensus guidelines on intrapartum fetal monitoring: Cardiotocography. Int J Gynaecol Obstet. 2015 Oct;131(1):13–24.
Li J, Chen ZZ, Huang L, Fang M, Li B, Fu X, Wang H. Automatic classification of fetal heart rate based on convolutional neural network. IEEE Internet of Things Journal 2019;6(2):1394–1401.
Petrozziello A, Jordanov I, Aris Papageorghiou T, Christopher Redman WG, Georgieva A. Deep learning for continuous electronic fetal monitoring in labor. Annu Int Conf IEEE Eng Med Biol Soc 2018:5866–5869.
Tang H, Wang T, Li M, Yang X. The design and implementation of cardiotocography signals classification algorithm based on neural network. Comput Math Methods Med 2018;8568617.
Ma’sum MA, Riskyana Dewi Intan P, Jatmiko W, Krisnadhi AA, Setiawan NA, Suarjaya IMAD. Improving deep learning classifier for fetus hypoxia detection in cardiotocography signal. International Workshop on Big Data and Information Security 2019:51–56.
Zhao Z, Zhang Y, Comert Z, Deng Y. Computer-aided diagnosis system of fetal hypoxia incorporating recurrence plot with convolutional neural network. Front Physiol 2019;10:255.
Zhao Z, Deng Y, Zhang Y, Zhang Y, Zhang X, Shao L. DeepFHR: intelligent prediction of fetal acidemia using fetal heart rate signals based on convolutional neural network. BMC Med Inform Decis Mak 2019;19(1):286.
Delay UH, et al. Non invasive wearable device for fetal movement detection. IEEE 15th International Conference on Industrial and Information Systems 2020:285–290.
Fotiadou E, van Sloun RJG, van Laar JOEH, Vullings R. A dilated inception CNN-LSTM network for fetal heart rate estimation. Physiol Meas 2021;42(4). https://doi.org/10.1088/1361-6579/abf7db.
Liang S, Li Q. Automatic evaluation of fetal heart rate based on deep learning. 2nd Information Communication Technologies Conference 2021:235–240.
Liu M, Lu Y, Long S, Bai J, Lian W, An attention-based CNN-BiLSTM hybrid neural network enhanced with features of discrete wavelet transformation for fetal acidosis classification. Expert Systems with Applications 2021;186:115714.
Baghel N, Burget R, Dutta MK. 1D-FHRNet: automatic diagnosis of fetal acidosis from fetal heart rate signals, Biomedical Signal Processing and Control 2022;71:102794.
Riskyana Dewi Intan P, Ma’sum MA, Alhamidi MR, Kurnianingsih, Jatmiko W. Generative adversarial networks for unbalanced fetal heart rate signal classification. ICT Express 2022;8(2):239–243.
He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. arXiv:1512.03385 [cs.CV]. Available online: https://arxiv.org/abs/1512.03385 (accessed on 1 December 2021).
TensorFlow Lite. Model Optimization. Available online: https://www.tensorflow.org/lite/performance/model_optimization (accessed on 21 October 2021).
Cheng, Y.; Wang, D.; Zhou, P.; Zhang, T. A Survey of Model Compression and Acceleration for Deep Neural Networks. arXiv:1710.09282 [cs.LG]. Available online: https://arxiv.org/abs/1710.09282 (accessed on 1 December 2021).
Lee, Y.J.; Moon, Y.H.; Park, J.Y.; Min, O.G. Recent R&D trends for lightweight deep learning. Electron. Telecommun. Trends 2019, 34, 40–50. https://doi.org/10.22648/ETRI.2019.J.340205.
Han, S.; Mao, H.; Dally, W.J. Deep Compression: Compressing Deep Neural Networks with Pruning, Trained Quantization and Huffman Coding. arXiv:1510.00149 [cs.CV]. Available online: https://arxiv.org/abs/1510.00149 (accessed on 1 December 2021).
Rastegari, M.; Ordonez, V.; Redmon, J.; Farhadi, A. XNOR-Net: ImageNet Classification Using Binary Convolutional Neural Networks. arXiv:1603.05279 [cs.CV]. Available online: https://arxiv.org/abs/1603.05279 (accessed on 1 December 2021).
Ullrich, K.; Meeds, E.; Welling, M. Soft Weight-Sharing for Neural Network Compression. arXiv:1702.04008 [stat.ML]. Available online: https://arxiv.org/abs/1702.04008 (accessed on 1 December 2021).
Hinton, G.; Vinyals, O.; Dean, J. Distilling the Knowledge in a Neural Network. arXiv:1503.02531 [stat.ML]. Available online: https://arxiv.org/abs/1503.02531 (accessed on 1 December 2021).
Lee KS, Park HJ, Kim JE, Kim HJ, Chon S, Kim S, Jang J, Kim JK, Jang S, Gil Y, Son HS. Compressed Deep Learning to Classify Arrhythmia in an Embedded Wearable Device. Sensors (Basel). 2022 Feb 24;22(5):1776.
Acknowledgements
None.
Funding
This work was supported by (1) the (IITP) Institute of Information & Communications Technology Planning & Evaluation grant funded by the Ministry of Science and ICT (No. IITP2020-0-00423), (2) the PACEN (Patient-Centered Clinical Research Coordinating Center) grant funded by the Ministry of Health & Welfare (No. HC22C0022) and (3) Korea Health Industry Development Institute grants funded by the Ministry of Health & Welfare (HI22C1302 (Korea Health Technology R&D Project)), Republic of Korea. The funder had no role in the design of the study, the collection, analysis and interpretation of the data and the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
K.S.L., E.S.C. and S.C.H. designed the study. K.S.L., E.S.C., Y.J.N., N.W.L., Y.S.Y., H.Y.K., K.H.A. and S.C.H. collected, analysed and interpreted the data. K.S.L., E.S.C. and S.C.H. wrote and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This retrospective study was approved by the Institutional Review Board (IRB) of Korea University Anam Hospital on November 8, 2021 (2021AN0324). Informed consent was waived by the IRB.
Consent for Publication
Not applicable.
Competing Interests
The authors have no competing interest to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, KS., Choi, E.S., Nam, Y.J. et al. Real-time Classification of Fetal Status Based on Deep Learning and Cardiotocography Data. J Med Syst 47, 82 (2023). https://doi.org/10.1007/s10916-023-01960-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-023-01960-1