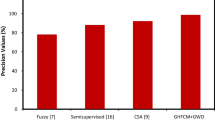
Abstract
Gene selection is imperative to clustering in light of gene articulation information, as a result of high Clustering quality. Clustering gene articulation information is a vital research subject in bioinformatics on the grounds that knowing which genes act correspondingly can prompt the disclosure of vital natural data. Many clustering systems have been proposed to the examination of gene articulation information got from microarray innovation. Clustering is one of the major procedures of investigating gene articulation information, fundamentally by contrasting gene articulation profiles or test articulation profiles. The Proposed strategy is an Agglo-Hi clustering algorithm which is accounted for the fuse of vicinity similarity estimates like Euclidean Distance, Manhattan Distance Chebyshev Distance, and Cosine Similarity for their execution. The technique is quality articulation information in microarray which is extricated and quality can be chosen from the preprocessed information, at that point the Agglo-Hi Clustering algorithm is utilized for quality information. The grouped information get approved utilizing legitimacy file and the outcome is gotten in light of nearness measures. To refine quality articulation information onto enhanced bunch quality by accelerating Unsupervised Learning stage and the execution of Agglo-Hi algorithm figures the Clustering quality, exactness and time unpredictability.







Similar content being viewed by others
References
Bala Subramaniyan R, Hullermeier E, Weskamp N, Kamper J (2004) Clustering of gene expression data using a local shape-based similarity measure. Bioinformatics © Oxford University Press
BalaAnand M, Karthikeyan N, Karthik S (2018) Designing a framework for communal software: based on the assessment using relation modelling. Int J Parallel Prog. https://doi.org/10.1007/s10766-018-0598-2
Boeva V, Tsiporkova E (2010) A multi-purpose time series data standardization method, intelligent systems: from theory to practice. Springer-Verlag Berlin Heidelberg, SCI 299: 445–460
Borg A, Lavesson N, Boeva V (2013) Comparison of clustering approaches for gene expression data. In: Jaeger M et al. (Eds.) Twelfth Scandinavian Conference on Artificial Intelligence. IOS Press
Bryan J (2004) Problems in gene clustering based on gene expression data. J Multivar Anal 90:44–66
Chalise P, Koestler DC, Bimali M, Yu Q, Fridley BL (2014) Integrative clustering methods for high-dimensional molecular data. Integrative Clustering Methods for High-Dimensional Molecular Data 3(3)
Chan EY, Ching WK, Ng MK, Huang JZ (2004) An optimization algorithm for clustering using weighted dissimilarity measures. Pattern Recogn 37(5):943–952
Chen H, Zhang Y, Gutman I (2016) A kernel-based clustering method for gene selection with gene expression data. J Biomed Inform 62:12–20
Costa IG, de A.T. de Carvalho F, de Souto MCP (2004) Comparative analysis of clustering methods for gene expression time course data. Genet Mol Biol 27(4):623–631
Jiang D, Tang C, Zhang A (2004) Cluster analysis for gene expression data: a survey. IEEE Trans Knowl Data Eng 16(11):1370–1386
Kerr G, Ruskin HJ, Crane M, Doolan P (2008) Techniques for clustering gene expression data. Comput Biol Med 38:283–293
Kormaksson M, Booth JG, Figueroa ME et al (2012) Integrative model-based clustering of microarray methylation and expression data. Ann Appl Stat 6:1327–1347
Liu J, Mohammed J, Carter J, Ranka S, Kahveci T, Baudis M (2006) Distance-based clustering of CGH data. Bioinformatics 22(16):1971–1978. https://doi.org/10.1093/bioinformatics/btl185
Mahima KM, Govindaraj M (2015) An effective validation methodology of proximity measures for clustering gene expression microarray data. International Journal of Innovative Research in Computer and Communication Engineering 3(2)
Makolo A, Adigun T (2016) Optimization of clustering algorithms for gene expression data analysis using distance measures. Int J Comput Appl 139(13)
McNicholas PD, Murphy TB (2010) Model-based clustering of microarray expression data via latent Gaussian mixture models. Bioinformatics 26:2705–2712
Moller-Levet C, Cho KH, Yin H, Wolkenhauer O (2003) Clustering of gene expression time-series data, Technical Report
Pirim H, Ekşioğlu B, Perkins A, Yüceer C (2012) Clustering of high throughput gene expression data. Comput Oper Res 39(12):3046–3061. https://doi.org/10.1016/j.cor.2012.03.008
Romdhane LB, Shili H, Ayeb B (2009) Mining microarray gene expression data with unsupervised possibilistic clustering and proximity graphs. Springer Science Business Media, LLC
Sarmah S, Bhattacharyya DK (2010) An effective technique for clustering incremental gene expression data. International Journal of Computer Science Issues 7(3):3
Seal S, Komarina S, Aluru S (2005) An optimal hierarchical clustering algorithm for gene expression data. Inf Process Lett 93:143–147
Visvanathan M, Adagarla BS, Gerald HL, Smith P (2009) Cluster validation: an integrative method for cluster analysis. IEEE International Conference on Bioinformatics and Biomedicine Workshop
Yeung KY, Medvedovic M, Bumgarner RE (2003) Clustering gene-expression data with repeated measurements. Genome Biol 4(5):R34
Zareizadeh Z, Helfroush MS, Rahideh A, Kazemi K (2018) A robust gene clustering algorithm based on clonal selection in multiobjective optimization framework. Expert Systems with Applications
Zhang W, Zhao D, Wang X (2013) Agglomerative clustering via maximum incremental path integral. Pattern Recogn 46:3056–3065
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kavitha, E., Tamilarasan, R. AGGLO-Hi clustering algorithm for gene expression micro array data using proximity measures. Multimed Tools Appl 79, 9003–9017 (2020). https://doi.org/10.1007/s11042-018-7112-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-7112-0