Abstract
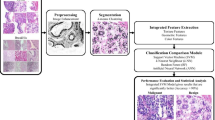
Breast cancer has become an important factor affecting human health. Diagnosis based on pathological images is considered the gold standard in the clinic. In this paper, an automatic breast cancer detection method based on hybrid features is proposed for pathological images. To obtain better segmentation results under conditions of crowded and chromatin-sparse nuclei, a 3-output convolutional neural network (CNN) is employed to segment the nuclei. Due to the weak correlation between the hematoxylin (H) and eosin (E) channels, texture features are separately extracted for the two channels, which provides more representative results. From multiple perspectives, the morphological features, spatial structural features and texture features are extracted and fused. Using a support vector machine (SVM) classifier with improved generalization, the pathological image is classified as benign or malignant on the basis of the relief method for feature selection. For the University of California, Santa Barbara database (UCSB), the classification accuracy of the method is 96.7%, and the area under the curve (AUC) is 0.983. The experimental results show that the proposed method yields superior classification performance compared with existing techniques.













Similar content being viewed by others
Change history
09 April 2019
The article Breast cancer classification in pathological images based on hybrid features, written by Cuiru Yu, Houjin Chen, Yanfeng Li, Yahui Peng, Jupeng Li and Fan Yang, was originally published electronically on the publisher’s internet portal (SpringerLink) on March 16, 2019 with open access.
References
Ferlay J, Soerjomataram I, Ervik M et al (2015) Breast cancer statistics. World Cancer Research Fund International. https://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/breast-cancer-statistics. Accessed 16 January 2015
Veta M, van Diest PJ, Kornegoor R et al (2013) Automatic nuclei segmentation in h&e stained breast cancer histopathology images. PLoS One 8(7):e70221
Petushi S, Garcia FU, Haber MM et al (2006) Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med Imaging 6(1):1–11
Vink JP, Van Leeuwen MB, Van Deurzen CHM et al (2013) Efficient nucleus detector in histopathology images. J Microsc 249(2):124–135
Sabeena BK, Nair MS, Bindu GR (2014) Automatic segmentation and classification of mitotic cell nuclei in histopathology images based on Active Contour Model. International Conference on Contemporary Computing and Informatics on IEEE, pp 740–744
Qi X, Xing F, Foran DJ et al (2012) Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans Biomed Eng 59(3):754
Zucker SW (1976) Region growing: childhood and adolescence. Computer Graphics & Image Processing 5(3):382–399
Ali S, Madabhushi A (2012) An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging 31(7):1448–1460
Sabeena BK, Nair MS, Bindu GR (2016) Automatic segmentation of cell nuclei using krill herd optimization based multi-thresholding and localized active contour model. Biocybernetics & Biomedical Engineering 36(4):584–596
Kong H, Gurcan M, Belkacem-Boussaid K (2011) Partitioning histopathological images: an integrated framework for supervised color-texture segmentation and cell splitting. IEEE Trans Med Imaging 30(9):1661
Plissiti ME, Nikou C (2012) Overlapping cell nuclei segmentation using a spatially adaptive active physical model. IEEE Transactions on Image Processing A Publication of the IEEE Signal Processing Society 21(11):4568
Chang H, Han J, Borowsky A et al (2013) Invariant delineation of nuclear architecture in glioblastoma multiforme for clinical and molecular association. IEEE Trans Med Imaging 32(4):670–682
Zhang M, Wu T, Bennett K (2015) Small blob identification in medical images using regional features from optimum scale. IEEE Trans Biomed Eng 62(4):1051–1062
Hatipoglu N, Bilgin G (2017) Cell segmentation in histopathological images with deep learning algorithms by utilizing spatial relationships. Med Biol Eng Comput 55(10):1–20
Kumar N, Verma R, Sharma S et al (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36(7):1550–1560
Anuranjeeta A, Shukla KK, Tiwari A et al (2017) Classification of histopathological images of breast cancerous and non cancerous cells based on morphological features. Biomedical & Pharmacology Journal 10(1):353–366
Doyle S, Agner S, Madabhushi A et al (2008) Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. IEEE International Symposium on Biomedical Imaging: From Nano To Macro 29:496–499
Hu Z, Tang J, Wang Z et al (2018) Deep learning for image-based cancer detection and diagnosis - A survey. Pattern Recogn 83(2018):134–149
Wu G, Lin Z, Han J, et al (2018) Unsupervised Deep Hashing via Binary Latent Factor Models for Large-scale Cross-modal Retrieval. Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, pp 2854–2860
Spanhol FA, Oliveira LS, Petitjean C et al (2016) Breast cancer histopathological image classification using Convolutional Neural Networks. International Joint Conference on Neural Networks, Vancouver
Spanhol F, Oliveira L, Petitjean C et al (2016) A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans Biomed Eng 63(7):1455–1462
Bayramoglu N , Kannala J, Heikkilä J (2017) Deep learning for magnification independent breast cancer histopathology image classification. International Conference on Pattern Recognition IEEE, pp 2440–2445
Spanhol F, Cavalin P, Oliveira L S, et al (2017) Deep Features for Breast Cancer Histopathological Image Classification. 2017 IEEE International Conference on Systems, Man and Cybernetics (SMC)
Gelasca ED, Byun J, Obara B, et al (2008) Evaluation and benchmark for biological image segmentation. IEEE International Conference on Image Processing, pp 1816–1819
Li X, Plataniotis KN (2015) Color model comparative analysis for breast cancer diagnosis using H and E stained images. SPIE Medical Imaging. International Society for Optics and Photonics 9420:94200L–94200L-6
He X, Han Z, Wei B (2000) Breast cancer histopathological image auto-classification using deep learning. Computer Engineering and Applications:1–000
Xiangsheng Z, Hong B, Chengquan Z (2014) Pathological diagnosis and differential diagnosis of breast. People’s Medical Publishing House, China
Okabe A, Boots B, Sugihara K et al (2001) Spatial Tessellations: Concepts and Applications of Voronoi Diagrams. John Wiley & Sons, Inc 3(4):357–363
De Berg M, Van Kreveld M, Overmars M et al (2000) Computational geometry: algorithms and applications. Math Gaz 19(3):333–334
Ruifrok AC, Johnston DA (2001) Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23(4):291–299
Bay H, Ess A, Tuytelaars T et al (2008) Speeded-up robust features (surf). Computer Vision & Image Understanding 110(3):346–359
Zulpe NS, Pawar VP (2012) Glcm textural features for brain tumor classification. International Journal of Computer Science Issues 9(3):354–359
Haraclick RM (1973) Texture Features for Image Classification. IEEE Trans Syst Man Cybern 3(6):610–621
Tahir MA, Bouridane A, Kurugollu F (2005) An fpga based coprocessor for glcm and haralick texture features and their application in prostate cancer classification. Analog Integr Circ Sig Process 43(2):205–215
Liu X (2014) Mass classification in mammogram with semi-supervised relief based feature selection. International Conference on Graphic and Image Processing, International Society for Optics and Photonics 9069:361–368
Collobert R, Bengio S, Marithoz J (2002) Torch: A Modular Machine Learning Software Library. Idiap, Switzerland
Otsu NA (1979) Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Veta M, Huisman A, Viergever M A, et al (2011) Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images. IEEE International Symposium on Biomedical Imaging: From Nano To Macro, pp 618–621
Lei T, Jia X, Zhang Y et al (2018) Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering. IEEE Trans Fuzzy Syst PP(99):1
Argandacarreras I, Kaynig V, Rueden C et al (2017) Trainable weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33(15):2424
Song Y, Li Q, Huang H, et al (2016) Histopathology Image Categorization with Discriminative Dimension Reduction of Fisher Vectors. Computer Vision - ECCV 2016 Workshops, Springer International Publishing, pp 306–317
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under no. 61872030 and no. 61571036.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised due to retrospective open access.
Rights and permissions
About this article
Cite this article
Yu, C., Chen, H., Li, Y. et al. Breast cancer classification in pathological images based on hybrid features. Multimed Tools Appl 78, 21325–21345 (2019). https://doi.org/10.1007/s11042-019-7468-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-7468-9