Abstract
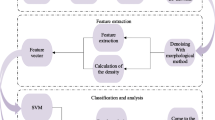
Breast density is known as a significant indicator of breast cancer risk prediction and greatly reduces the digital mammograms sensitivity. In this work, based on the simple pulse coupled neural network (SPCNN), a novel Morph_SPCNN model is proposed for dealing with the limitations of over-segmentation that commonly existed in density segmentation of mammograms. To evaluate the proposed model, the segmentation result is employed as a feature map of the breast density classification system. In addtion, the texture features of mammogram calculated based on the gray level co-occurrence matrix (GLCM) and the statistical features (mean, skewness, kurtosis) are extracted and input to the support vector machine (SVM) for breast density classification. Finally, the performance of SVM classifier is evaluated based on the ten-fold cross-validation. Our method is verified both on the MIAS dataset, DDSM database and hybrid dataset (MIAS database and Gansu Provincial Academy of Medical Sciences (GPAMS) database), respectively achieving 87.80%, 94.89% and 95.37% accuracy for breast density classification. The experimental results indicate that our proposed method has greatly improved the performance of breast density segmentation and classification.













Similar content being viewed by others
References
AC Society (2019) Cancer facts and figures 2019 https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2019.html
AC of Radiology, D’Orsi CJ et al (2013) ACR BI-RADS atlas: breast imaging reporting and data system; mammography, ultrasound, magnetic resonance imaging, follow-up and outcome monitoring, data dictionary, ACR American College of Radiology
Anguita D, Ridella S, Rivieccio F (2005) K-fold generalization capability assessment for support vector classifiers. In: Neural networks, 2005. IJCNN ’05. Proceedings. 2005 IEEE international joint conference on. https://doi.org/10.1109/IJCNN.2005.1555964
Angulo J (2016) Generalised morphological image diffusion. Nonlinear Anal Theory Methods Appl 134:1–30. https://doi.org/10.1016/j.na.2015.12.015
B.C. U.K. (2019) Key facts about breast cancer http://www.breastcanceruk.org.uk/
Blot L, Zwiggelaar R (2001) Background texture extraction for the classification of mammographic parenchymal patterns. In: MIUA, pp 145–148
Bosch A, Munoz X, Oliver A, Marti J (2006) Modeling and classifying breast tissue density in mammograms. 2:1552–1558. https://doi.org/10.1109/CVPR.2006.188
Bowyer K, Kopans D, Kegelmeyer W, Moore R, Sallam M, Chang K, Woods K (1996) The digital database for screening mammography. In: Third international workshop on digital mammography, vol 58, p 27
Chang C-C, Lin C-J (2011) Libsvm: A library for support vector machines. ACM Trans Intell Syst Technol. https://doi.org/10.1145/1961189.1961199
Chen Z, Denton ERE, Zwiggelaar R (2011) Local feature based mammographic tissue pattern modelling and breast density classification. 1:351–355. https://doi.org/10.1109/BMEI.2011.6098279
Chen Y, Park S, Ma Y, Ala R (2011) A new automatic parameter setting method of a simplified pcnn for image segmentation. IEEE Trans Neural Netw 22(6):880–892. https://doi.org/10.1109/TNN.2011.2128880
Chen Y, Tao J, Liu L, Xiong J, Xia R, Xie J, Zhang Q, Yang K (2020) Research of improving semantic image segmentation based on a feature fusion model. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02066-z
Chen Y, Wang J, Chen X, Zhu M, Yang K, Wang Z, Xia R (2019) Single-image super-resolution algorithm based on structural self-similarity and deformation block features. IEEE Access 7:58791–58801. https://doi.org/10.1109/ACCESS.2019.2911892
Chen Y, Wang J, Liu S, Chen X, Xiong J, Xie J, Yang K (2019) Multiscale fast correlation filtering tracking algorithm based on a feature fusion model. Concurr Comput Pract Exp e5533. https://doi.org/10.1002/cpe.5533
Chen Y, Xiong J, Xu W, Zuo J (2019) A novel online incremental and decremental learning algorithm based on variable support vector machine. Clust Comput 22(3):7435–7445. https://doi.org/10.1007/s10586-018-1772-4
Chen Y, Xu W, Zuo J, Yang K (2019) The fire recognition algorithm using dynamic feature fusion and iv-svm classifier. Clust Comput 22 (3):7665–7675. https://doi.org/10.1007/s10586-018-2368-8
Chen Y, Tao J, Zhang Q, Yang K, Chen X, Xiong J, Xia R, Xie J Saliency detection via the improved hierarchical principal component analysis method. Wirel Commun Mob Comput (Online) 2020. https://doi.org/10.1155/2020/8822777
Cortes C, Vapnik V (1995) Support-vector networks, vol 20. https://doi.org/10.1023/A:1022627411411
Deng J, Ma Y, Deng-ao L, Zhao J, Liu Y, Zhang H (2020) Classification of breast density categories based on se-attention neural networks. Comput Methods Programs Biomed 105489. https://doi.org/10.1016/j.cmpb.2020.105489
Deng XY, Yi-De MA (2012) Pcnn model automatic parameters determination and its modified model. Acta Electronica Sinica 5(5):955–964. https://doi.org/10.3969/j.issn.0372-2112.2012.05.015
Eckhorn R, Reitboeck H, Arndt M, Dicke P (1990) Feature linking via synchronization among distributed assemblies: simulations of results from cat visual cortex. Neural Comput 2(3):293–307. https://doi.org/10.1162/neco.1990.2.3.293
Ekblad U, Kinser JM, Atmer J, Zetterlund N (2004) The intersecting cortical model in image processing. Nucl Instrum Methods Phys Res B 525(1):392–396. https://doi.org/10.1016/j.nima.2004.03.102
Elshinawy MY, Badawy AHA, Abdelmageed WW, Chouikha MF (2011) Effect of breast density in selecting features for normal mammogram detection. In: IEEE International symposium on biomedical imaging: from nano to macro. https://doi.org/10.1109/ISBI.2011.5872374
Eng A, Gallant Z, Shepherd J, Mccormack V, Li J, Dowsett M, et al. (2014) Digital mammographic density and breast cancer risk:a case-control study of six alternative density assessment methods. Breast Cancer Res 16:439. https://doi.org/10.1186/s13058-014-0439-1
Gong X, Yang Z, Wang D, Qi Y, Ma Y (2019) Breast density analysis based on glandular tissue segmentation and mixed feature extraction. Multimed Tools Appl 78(5):31185–31214. https://doi.org/10.1007/s11042-019-07917-2
Gu X (2008) Feature extraction using unit-linking pulse coupled neural network and its applications. Neural Process Lett 27(1):25–41. https://doi.org/10.1007/s11063-007-9057-6
Hage IS, Hamade RF (2013) Segmentation of histology slides of cortical bone using pulse coupled neural networks optimized by particle-swarm optimization. Comput Med Imaging Graph 37(7-8). https://doi.org/10.1016/j.compmedimag.2013.08.003
Hamidinekoo A, Denton E, Rampun A, Honnor K, Zwiggelaar R (2018) Deep learning in mammography and breast histology, an overview and future trends. Med Image Anal 35:303–312. https://doi.org/10.1016/j.media.2018.03.006
Hassanien AE, Kim TH (2012) Breast cancer mri diagnosis approach using support vector machine and pulse coupled neural networks. J Appl Log 10(4):277–284. https://doi.org/10.1016/j.jal.2012.07.003
He W, Denton ERE, Stafford K, Zwiggelaar R (2011) Mammographic image segmentation and risk classification based on mammographic parenchymal patterns and geometric moments. Biomed Signal Process Control 6(3):321–329. https://doi.org/10.1016/j.bspc.2011.03.008
Holzinger A (2016) Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inform 3(2):119–131
Holzinger A, Carrington AM, Muller H (2020) Measuring the quality of explanations: The system causability scale (scs): comparing human and machine explanations. Künstliche Intelligenz 1–6
Holzinger A, Kieseberg P, Weippl E, Tjoa AM (2018) Current advances, trends and challenges of machine learning and knowledge extraction: from machine learning to explainable ai. https://doi.org/10.1007/978-3-319-99740-7_1
Holzinger A, Langs G, Denk H, Zatloukal K, Muller H (2019) Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip Rev Data Min Knowl Discov 9(4). https://doi.org/10.1002/widm.1312
Hsu C, Lin C (2002) A comparison of methods for multiclass support vector machines. IEEE Trans Neural Netw 13(2):415–425. https://doi.org/10.1109/72.991427
Jia T, Zhang H, Bai YK (2015) Benign and malignant lung nodule classification based on deep learning feature. J Med Imaging Health Inform 5(8):1936–1940. https://doi.org/10.1166/jmihi.2015.1673
Johnson JL, Ritter D (1993) Observation of periodic waves in a pulse-coupled neuralnetwork. Opt Lett 18(15):1253–5. https://doi.org/10.1364/OL.18.001253
Kinser JM (1996) Simplified pulse-coupled neural network. Proc SPIE 2760:563–567. https://doi.org/10.1117/12.235951
Kumar I, Bhadauria HS, Virmani J (2015) Wavelet packet texture descriptors based four-class birads breast tissue density classification. Procedia Comput Sci 70:76–84. https://doi.org/10.1016/j.procs.2015.10.042
Kumar I, Bhadauria HS, Virmani J, Thakur S (2017) A classification framework for prediction of breast density using an ensemble of neural network classifiers. Biocybern Biomed Eng 37(1):217–228. https://doi.org/10.1016/j.bbe.2017.01.001
Kuntimad G, Ranganath HS (1999) Perfect image segmentation using pulse coupled neural networks. IEEE Trans Neural Netw 10(3):591–598. https://doi.org/10.1109/72.761716
Lian J, Li K (2020) A review of breast density implications and breast cancer screening. Clin Breast Cancer 12:30–38. https://doi.org/10.1016/j.clbc.2020.03.004
Lian J, Yang Z, Sun W, Guo Y, Zheng L, Li J, et al. (2019) An image segmentation method of a modified spcnn based on human visual system in medical images. Neurocomputing 333:292–306. https://doi.org/10.1016/j.neucom.2018.12.007
Liao Z, Zhang R, He S, Zeng D, Wang J, Kim H-J (2019) Deep learning-based data storage for low latency in data center networks. IEEE Access 7:26411–26417. https://doi.org/10.1109/ACCESS.2019.2901742
Lu X, Wang W, Ma C, Shen J, Shao L, Porikli F (2019) See more, know more: Unsupervised video object segmentation with co-attention siamese networks. In: 2019 IEEE/CVF conference on Computer Vision and Pattern Recognition (CVPR), pp 3618–3627. https://doi.org/10.1109/CVPR.2019.00374
Lu X, Wang W, Shen J, Tai Y, Crandall DJ, Hoi SCH (2020) Learning video object segmentation from unlabeled videos. Comput Vis Pattern Recognit
Ma Y-D, Dai R-L, Li L (2002) Automated image segmentation using pulse coupled neural networks and image’s entropy. China Inst Commun 23 (1):46–51
Ma Y, Dai R, Li L, Wei L (2002) Image segmentation of embryonic plant cell using pulse-coupled neural networks. Chin Sci Bull 47:169–173. https://doi.org/10.1360/02tb9040
Machida Y, Tozaki M, andss Tamiko Yoshida AS (2015) Breast density: the trend in breast cancer screening. Breast Cancer 22(3):253–261. https://doi.org/10.1007/s12282-015-0602-2
Malkov S, Shepherd JA, Scott CG, Tamimi RM, Ma L, et al. (2016) Mammographic texture and risk of breast cancer by tumor type and estrogen receptor status. Breast Cancer Res 18(1):122. https://doi.org/10.1186/s13058-016-0778-1
Manduca A, Carston MJ, Heine JJ, Scott CG, Pankratz VS, Brandt KR, et al. (2009) Texture features from mammographic images and risk of breast cancer, Cancer Epidemiology. Biomarkers and Prevention 18(3):837–845. https://doi.org/10.1158/1055-9965.EPI-08-0631
Masci J, Angulo J, Schmidhuber J (2013) A learning framework for morphological operators using counter–harmonic mean. 7883:329–340. https://doi.org/10.1007/978-3-642-38294-9_28
McCormack AV (2006) Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev 15 (6):1159–1169. https://doi.org/10.1158/1055-9965.EPI-06-0034
Mellouli D, Hamdani TM, Sanchez-Medina JJ, Ayed MB, Alimi AM (2019) Morphological convolutional neural network architecture for digit recognition. IEEE Trans Neural Netw Learn Syst 1–10. https://doi.org/10.1109/TNNLS.2018.2890334
Moon WK, Chang JF, Lo CM, Chang JM, Lee SH, Shin SU, et al. (2018) Quantitative breast density analysis using tomosynthesis and comparison with mri and digital mammography. Comput Methods Programs Biomed 154:99–107. https://doi.org/10.1016/j.cmpb.2017.11.008
Muhimmah I (2006)
Muštra M, Grgić M, Delač K (2010) Feature selection for automatic breast density classification. In: International symposium elmar
Oliver A, Freixenet J, Marti R, Pont J, Perez E, Denton ERE, Zwiggelaar R (2008) A novel breast tissue density classification methodology 12(1):55–65. https://doi.org/10.1109/TITB.2007.903514
Oliver A, Freixenet J, Martí R, Pont J, Pérez E, Denton ERE, Zwiggelaar R (2008) A novel breast tissue density classification methodology. IEEE Trans Inf Technol Biomed 12(1):55–65. https://doi.org/10.1109/TITB.2007.903514
Oliver A, Freixenet J, Zwiggelaar R (2005). https://doi.org/10.1109/ICIP.2005.1530291
Oliver A, Tortajada M, Lladó X, Freixenet J, Ganau S, Tortajada L, et al. (2015) Breast density analysis using an automatic density segmentation algorithm. J Digit Imaging 28:604–612. https://doi.org/10.1007/s10278-015-9777-5
Otsu N (2007) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern B Cybern 9(1):62–66. https://doi.org/10.1109/TSMC.1979.4310076
Parthalain NM, Jensen R, Shen Q, Zwiggelaar R (2010) Fuzzy-rough approaches for mammographic risk analysis. 14(2):225–244. https://doi.org/10.3233/IDA-2010-0418
Petroudi S, Constantinou I, Tziakouri C, Pattichis MS, Pattichis CS (2013) Investigation of am-fm methods for mammographic breast density classification. 1–4. https://doi.org/10.1109/BIBE.2013.6701633
Petroudi S, Kadir T, Brady M (2003). https://doi.org/10.1109/IEMBS.2003.1279885
Rampun A, Morrow P, Scotney B, Winder J (2017) Breast density classification using multiresolution local quinary patterns in mammograms. In: Conference on medical image understanding and analysis. https://doi.org/10.1007/978-3-319-60964-5_32
Rampun A, Scotney BW, Morrow PJ, Wang H (2019) Breast density classification using local septenary patterns: a multi-resolution and multi-topology approach. In: 2019 IEEE 32nd international symposium on computer-based medical systems (CBMS). https://doi.org/10.1109/CBMS.2019.00133
Ranganath HS, Kuntimad G (1996) Iterative segmentation using pulse-coupled neural networks. Proc SPIE Int Soc Opt En 2760:543–554. https://doi.org/10.1117/12.235943
Remes V, Haindl M (2015) Classification of breast density in x-ray mammography. In: International workshop on computational intelligence for multimedia understanding. https://doi.org/10.1109/IWCIM.2015.7347085
Ribli D, Horváth A, Unger Z, Pollner P, Csabai I (2017) Detecting and classifying lesions in mammograms with deep learning. Sci Rep 8(1):1–7. https://doi.org/10.1038/s41598-018-22437-z
Shen D, Wu G, Suk H (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19(1):221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Strand F, Humphreys K, Cheddad A, Tornberg S, Azavedo E, Shepherd JA, Hall P, Czene K (2016) Novel mammographic image features differentiate between interval and screen-detected breast cancer: a case-case study. Breast Cancer Res 18(1):100. https://doi.org/10.1186/s13058-016-0761-x
Suckling J (1994) The mammographic image analysis society digital mammogram database. Digit Mammo 375–386
Surajudeen A, Reyer Z (2017) Breast density segmentation based on fusion of super pixels and watershed transform. Int J Comput Appl 161(12):1–7. https://doi.org/10.5120/ijca2017913208
Tzikopoulos S, Mavroforakis M, Georgiou HV, Dimitropoulos N, Theodoridis S (2011) A fully automated scheme for mammographic segmentation and classification based on breast density and asymmetry. Comput Methods Prog Biomed 102(1):47–63. https://doi.org/10.1016/j.cmpb.2010.11.016
Virmani J, Dey N, Kumar V et al (2016) Pca-pnn and pca-svm based cad systems for breast density classification, Springer. https://doi.org/10.1007/978-3-319-21212-8_7
Wang Y, Jiao J (2011) Detection of regions of interest from breast tumorul trasound images using improved pcnn. Opt.Precis.Eng 19(6). https://doi.org/10.3788/ope.20111906.1398
Wang J, Kato F, Yamashita H, Baba M, Cui Y, Li R, et al. (2017) Automatic estimation of volumetric breast density using artificial neural network-based calibration of full-field digital mammography: feasibility on japanese women with and without breast cancer. J Digit Imaging 30(2):215–227. https://doi.org/10.1007/s10278-016-9922-9
Williams CKI (2003) Learning with kernels: Support vector machines, regularization, optimization, and beyond. J Am Stat Assoc 98(462):489–489. https://doi.org/10.1198/jasa.2003.s269
Wolfe JN (1976) Risk for breast cancer development determined by mammographic parenchymal pattern. Cancer 37:2486–2492. https://doi.org/10.1002/1097-0142(197605)37:53.0.CO;2-8
Yang Z, Lian J, Li S, Guo Y, Qi Y, Ma Y (2018) Heterogeneous spcnn and its application in image segmentation. Neurocomputing 285 (3):196–203. https://doi.org/10.1016/j.neucom.2018.01.044
Zhan K, Shi J, Wang H, Xie Y, Li Q (2017) Computational mechanisms of pulse-coupled neural networks: a comprehensive review. Arch Comput Methods Eng 24(3):573–588. https://doi.org/10.1007/s11831-016-9182-3
Zhan K, Zhang H, Ma Y (2009) New spiking cortical model for invariant texture retrieval and image processing. IEEE Trans Neussral Netw 20 (12):1980–1986. https://doi.org/10.1109/TNN.2009.2030585
Acknowledgement
This work is jointly supported by the Natural Science Foundation of Gansu Province (No.18JR3RA288) and the Fundamental Research Funds for the Central Universities of China (No.lzujbky-2017-it72 and No.lzujbky-2018-it61).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, Y., Yang, Z., Lei, J. et al. Morph_SPCNN model and its application in breast density segmentation. Multimed Tools Appl 80, 2821–2845 (2021). https://doi.org/10.1007/s11042-020-09796-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09796-4