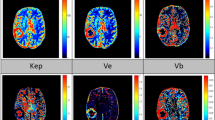
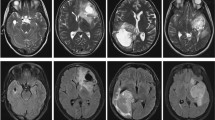
Abstract
Computer-aided diagnosis has attracted attention for the accurate grading of cerebral glioma. Most algorithms are only effective in relatively large datasets. Although multicenter data sharing is expanding, the results of cerebral glioma grading are not promising for multicenter data. Considering that multicenter images differ in contrast, we propose an effective image standardization method to reduce the disparity in image contrast of different datasets. The method is adopted in multiple sets of comparative experiments on a public dataset (BraTS2017) and a local dataset. The classification accuracy of experimental data relative to that of multicenter data without image normalization is improved by approximately 25% on average. Results demonstrate that the proposed approach is effective in solving the image contrast disparity of multicenter data. It also addresses the challenge of limited effective sample size in accurate cerebral glioma grading. The novel image standardization technology proposed in this work is a promising solution that can be integrated into expert systems.





Similar content being viewed by others
References
Abdelaziz Ismael SA, Mohammed A, Hefny H (Jan 2020) An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif Intell Med 102:101779
Ackaouy A, Courty N, Vallee E, Commowick O, Barillot C, Galassi F (2020) Unsupervised domain adaptation with optimal transport in multi-site segmentation of multiple sclerosis lesions from MRI data. Front Comput Neurosci 14:19
S. Bakas et al., "Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features," Sci Data, vol. 4, p. 170117, Sep 5 2017.
Bargshady G, Zhou X, Deo RC, Soar J, Whittaker F, Wang H (2020) Enhanced deep learning algorithm development to detect pain intensity from facial expression images. Expert Syst Appl 149:113305
Celik T (2012) Two-dimensional histogram equalization and contrast enhancement. Pattern Recogn 45(10):3810–3824
Chen SD, Ramli AR (2004) Minimum mean brightness error bi-histogram equalization in contrast enhancement. IEEE Trans Consum Electron 49(4):1310–1319
Chen X, Xie T, Fang J, Xue W, Tong H, Kang H, Wang S, Yang Y, Xu M, Zhang W (2017) Quantitative in vivo imaging of tissue factor expression in glioma using dynamic contrast-enhanced MRI derived parameters. Eur J Radiol 93:236–242
Chen X et al (2019) Automatic histogram specification for Glioma grading using Multicenter data. J Healthc Eng:9414937
Chen C, Dou Q, Chen H, Qin J, Heng PA (2020) Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation. IEEE Trans Med Imaging 39(7):2494–2505
B. H. Diplas et al., "Sensitive and rapid detection of TERT promoter and IDH mutations in diffuse gliomas," Neuro Oncol, vol. 21, no. 4, pp. 440–450, Mar 18 2019
Ghassemi N, Shoeibi A, Rouhani M (2020) Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control 57:101678
Graham RN, Perriss RW, Scarsbrook AF (2005) DICOM demystified: a review of digital file formats and their use in radiological practice. Clin Radiol 60(11):1133–1140
Han S, Liu Y, Cai SJ, Qian M, Ding J, Larion M, Gilbert MR, Yang C (2020) IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer 122(11):1580–1589
Jin H, Luo Y, Li P, Mathew J (2019) A review of secure and privacy-preserving medical data sharing. IEEE Access 7:61656–61669
Joseph J, Sivaraman J, Periyasamy R, Simi VR (2017) An objective method to identify optimum clip-limit and histogram specification of contrast limited adaptive histogram equalization for MR images. Biocybernet Biomed Eng 37(3):489–497
Kudulaiti N, Qiu T, Lu J, Zhang H, Zhang Z, Guan Y, Zhuang D, Wu J (2019) Combination of magnetic resonance spectroscopy and 11C-methionine positron emission tomography for the accurate diagnosis of non-enhancing Supratentorial Glioma. Korean J Radiol 20(6):967–975
Li W, Zhao Y, Chen X, Xiao Y, Qin Y (2019) Detecting Alzheimer's disease on small dataset: a knowledge transfer perspective. IEEE J Biomed Health Inform 23(3):1234–1242
Li L, Wang K, Ma X, Liu Z, Wang S, du J, Tian K, Zhou X, wei W, Sun K, Lin Y, Wu Z, Tian J (2019) Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur J Radiol 118:81–87
Liang D, Gao X, Lu W, He L (2020) Deep multi-label learning for image distortion identification. Signal Process 172:107536
Li-Chun Hsieh K, Chen CY, Lo CM (2017) Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med 83:102–108
Liu J, Chen F, Pan C, Zhu M, Zhang X, Zhang L, Liao H (2018) A cascaded deep convolutional neural network for joint segmentation and genotype prediction of brainstem Gliomas. IEEE Trans Biomed Eng 65(9):1943–1952
Liu C, Sui X, Kuang X, Liu Y, Gu G, Chen Q (2019) Optimized contrast enhancement for infrared images based on global and local histogram specification. Remote Sens 11(7):849
Liu Q, Jiang P, Jiang YH, Ge HJ, Li SL, Jin HW, Li YX (2019) Prediction of aneurysm stability using a machine learning model based on PyRadiomics-derived morphological features. Stroke 50(9):2314–2321
Liu M, Zhou Z, Shang P, Xu D (2020) Fuzzified image enhancement for deep learning in Iris recognition. IEEE Trans Fuzzy Syst 28(1):92–99
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of Tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Lu Z, Bai Y, Chen Y, Su C, Lu S, Zhan T, Hong X, Wang S (2020) The classification of gliomas based on a pyramid dilated convolution resnet model. Pattern Recogn Lett 133:173–179
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp C, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, van Leemput K (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024
Nyul LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imag 19(2):143
Patel V (2019) A framework for secure and decentralized sharing of medical imaging data via blockchain consensus. Health Informatics J 25(4):1398–1411
Perone CS, Ballester P, Barros RC, Cohen-Adad J (2019) Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. Neuroimage 194:1–11
Raab P (2010) Cerebral gliomas: diffusional kurtosis imaging analysis of microstructural differences. Radiology 254(3):876–881
Rao BS (2020) Dynamic histogram equalization for contrast enhancement for digital images. Appl Soft Comput 89:106114
Roy R, Ghosh S, Ghosh A (2020) Clinical ultrasound image standardization using histogram specification. Comput Biol Med 120:103746
Rundo L, Tangherloni A, Nobile MS, Militello C, Besozzi D, Mauri G, Cazzaniga P (2019) MedGA: a novel evolutionary method for image enhancement in medical imaging systems. Expert Syst Appl 119:387–399
Sen D, Pal SK (May 2011) Automatic exact histogram specification for contrast enhancement and visual system based quantitative evaluation. IEEE Trans Image Process 20(5):1211–1220
Sepp M (2007) High-quality two-stage resampling for 3-D volumes in medical imaging. Med Image Anal 11(4):346–360
van Griethuysen JJM et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107
Wachinger C, Reuter M (2016) Domain adaptation for Alzheimer's disease diagnostics. Neuroimage 139:470–479
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13(4):600–612
Wang R, Bao HB, du WZ, Chen XF, Liu HL, Han DY, Wang LG, Wu JN, Wang CL, Yang MC, Liu ZW, Zhang N, Teng L (Jan 2019) P68 RNA helicase promotes invasion of glioma cells through negatively regulating DUSP5. Cancer Sci 110(1):107–117
Xie T, Chen X, Fang J, Kang H, Xue W, Tong H, Cao P, Wang S, Yang Y, Zhang W (2018) Textural features of dynamic contrast-enhanced MRI derived model-free and model-based parameter maps in glioma grading. J Magn Reson Imaging 47(4):1099–1111
Xu G, Xu X, Wang X, Wang X (2019) Order-encoded quantum image model and parallel histogram specification. Quantum Information Process 18(11)
Yoo JC, Ahn CW (2012) Image matching using peak signal-to-noise ratio-based occlusion detection. IET Image Process 6(5):483
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, G., Bai, J., Yang, G. et al. Magnetic resonance imaging standardization for accurate grading of cerebral gliomas. Multimed Tools Appl 81, 41477–41496 (2022). https://doi.org/10.1007/s11042-020-10487-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-10487-3