Abstract
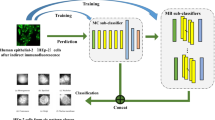
Manual analysis of the indirect-immunofluorescence (IIF) human epithelial cell Type-2 (HEp-2) cell image for the diagnosis of an auto-immune disease is a subjective and time-consuming process, and it is also prone to human-errors. The present work proposes an automatic capsule neural network (CapsNet) based framework for HEp-2 cell image classification to compensate for the deficiencies present in the prominent convolution neural network (CNN) based frameworks. In CNNs, the spatial relationship between the features present in the anti-nuclear antibodies (ANA) patterns, found in the IIF HEp-2 cell image (ANA-IIF image) is lost which increases the chance of detection of false-positives. In the proposed CapsNet based model, the max-pooling layer has been replaced with advanced dynamic routing algorithm and scalar outputs are replaced with the vector output, thus the richer representation of the same feature without the loss of spatial relationship with respect to the other features are made possible. The proposed framework recognizes ANA-IIF images with an average accuracy of 95.00% for 10-fold cross-validations. The experimental result also shows that the proposed model performs better than the other CNN based classification models for human epithelial cell image classification task.







Similar content being viewed by others
References
Afshar P, Mohammadi A, Plataniotis KN (2018) Brain tumor type classification via capsule networks, in 2018 25th IEEE International Conference on Image Processing (ICIP). IEEE, 3129–3133
American College ofRheumatology. Position statement:Methodology oftesting for antinuclear antibodies (2011) www.rheumatology.org/practice/clinical/position/ana_position_stmt.pdf. Accessed 13 Jan 2020
Baydilli YY, Atila U (2020) Classification of white blood cells using capsule networks. Comput Med Imaging Graph 80(101699):0895–6111. https://doi.org/10.1016/j.compmedimag.2020.101699
Bradwell AR, Hughes RG (2007) Atlas of Hep-2 patterns and laboratory techniques. Binding Site, 3rd ed
Capsule Networks Are Shaking up AI — Here’s How to Use Them (n.d.), https://hackernoon.com/capsule-networks-are-shaking-up-ai-heres-how-to-use-themc233a0971952. Accessed 18 Apr 2019
Cascio D, Taormina V, Raso G (2019) Deep CNN for IIF images classification in autoimmune diagnostics. Appl Sci 9(8):1618
Foggia P, Percannella G, Soda P, Vento M (2013) Benchmarking hep-2 cells classification meth- ods. IEEE Trans Med Imaging 32(10):1878–1889
Gao Z, Wang L, Zhou L, Zhang J (2017) HEp-2 cell image classification with deep convolutional neural networks. IEEE J Biomed Health Informatics 21(2):416–428. https://doi.org/10.1109/JBHI.2016.2526603
Glorot X, Bengio Y (2010) Understanding the difficulty of training deep feedforward neural networks. Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics 9:249–256
Hernández G, Zamora E, Sossa H, Téllez G, Furlán F (2020) Hybrid neural networks for big data classification. Neurocomputing 390:327–340,ISSN 0925–2312. https://doi.org/10.1016/j.neucom.2019.08.095
Hiemann R, Büttner T, Krieger T, Roggenbuck D, Sack U, Conrad K (2009) Challenges of automated screening and differentiation of non-organ specific autoantibodies on hep-2 cells. Autoimmun Rev 9(1):17–22
Iesmantas T, Alzbutas R (2018) Convolutional capsule network for classification of breast cancer histology images, International Conference Image Analysis and Recognition. Springer, New York, pp 853–860
LaLonde R, Bagci U (2018) Capsules for object segmentation. ArXiv, abs/1804.04241
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Lei H, Han T, Zhou F, Yu Z, Qin J, Elazab A, Lei B (2018) A deeply supervised residual network for HEp-2 cell classification via cross-modal transfer learning. Pattern Recogn 79:290–302
Lu M, Gao L, Guo X, Liu Q, Yin J (2017) HEp-2 cell image classification method based on very deep convolutional networks with small datasets, Proc. SPIE 10420, Ninth International Conference on Digital Image Processing (ICDIP 2017), 1042040
Majtner T, Baji’c B, Lindblad J, Sladoje N, Blanes-Vidal V, Nadimi ES (2019) On the effectiveness of generative adversarial Networks as HEp-2 image augmentation tool. Scandinavian Conference on Image Analysis, Springer, 439–451. https://doi.org/10.1007/978-3-030-20205-7_36
Mobiny A., Van Nguyen H (2018) Fast CapsNet for lung cancer screening. In: Frangi A, Schnabel J, Davatzikos C, Alberola-López C, Fichtinger G (eds) Medical image computing and computer assisted intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in computer Science, vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_82
Phan HTH, Kumar A, Kim J, Feng D (2016) Transfer learning of a convolutional neural network for HEp-2 cell image classification. IEEE 13th International Symposium on Biomedical Imaging (ISBI). Prague 2016:1208–1211. https://doi.org/10.1109/ISBI.2016.7493483
Qi X, Zhao G, Chen J (2016) Exploring illumination robust descriptors for human epithelial type 2 cell classification. Pattern Recogn 60:420–429. https://doi.org/10.1016/j.patcog.2016.05.032
Rodrigues LF, Naldi MC, Mari JF (2017) Exploiting convolutional neural networks and preprocessing techniques for HEp-2 cell classification in immunofluorescence images, 2017 30th SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Niteroi, pp 170–177. https://doi.org/10.1109/SIBGRAPI.2017.29
Rodrigues LF, Naldi MC and Mari JF (2017) HEp-2 Cell Image Classification Based on Convolutional Neural Networks, Workshop of Computer Vision (WVC), Natal, pp 13–18. https://doi.org/10.1109/WVC.2017.00010
Rodrigues LF, Naldi MC, Mari JF (2019) Comparing convolutional neural networks and preprocessing techniques for HEp-2 cell classification in immunofluorescence images. Comput Biol Med 116:103542. https://doi.org/10.1016/j.compbiomed.2019.103542
Sabour S, Frosst N, Hinton GE (2017) Dynamic routing between capsules. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS’17). Curran Associates Inc., Red Hook, NY, 3859–3869
Soda P (2007) Early experiences in the staining pattern classification of HEp-2 slides, Twentieth IEEE International Symposium on Computer-Based Medical Systems (CBMS’07), Maribor, pp 219–224. https://doi.org/10.1109/CBMS.2007.42
Vununu C, Lee S-H, Kwon K-R (2019) A deep feature extraction method for HEp-2 cell image classification. Electronics 8(1):20
Wiik AS, Høier-Madsen M, Forslid J, Charles P, Meyrowitsch J (2010) Anti- nuclear antibodies: a contemporary nomenclature using hep-2 cells. J Ofautoimmun 35(3):276–290
Wiliem A, Wong Y, Sanderson C, Hobson P, Chen S, Lovell BC (2013) Classification of Human Epithelial type 2 cell indirect immunofluoresence images via codebook based descriptors. In Proceedings of the 2013 IEEE Workshop on Applications of Computer Vision (WACV) (WACV’13). IEEE Computer Society, pp 95–102. https://doi.org/10.1109/WACV.2013.6475005
Xiang X, Xingkun W, Feng L (2017) Cellular image classification
Zhang X, Zhao SG (2019) Fluorescence microscopy image classification of 2D HeLa cells based on the CapsNet neural network. Med Biol Eng Comput 57(6):1187–1198. https://doi.org/10.1007/s11517-018-01946-z
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Maurya, R., Pathak, V.K. & Dutta, M.K. Computer-aided diagnosis of auto-immune disease using capsule neural network. Multimed Tools Appl 81, 13611–13632 (2022). https://doi.org/10.1007/s11042-021-10534-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-10534-7