Abstract
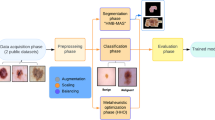
Melanoma is considered the deadliest form of skin cancer, and the number of cases is increasing day by day. The early diagnosis of melanoma is critical, as it significantly increases the patient’s chance of survival. However, distinguishing melanoma from other skin lesion types by the physician can be a complicated process due to the diversity of its structural and textural features. Numerous computer-aided diagnosis (CAD) systems have been developed to assist the physician in detecting melanoma during recent years. The segmentation is a critical step for CAD systems, as it directly contributes to the performance of both feature extraction and classification steps. The optimization of the hyperparameters of deep learning methods is a challenging research topic. In this paper, the Bayesian optimized SegNet approach is proposed for precise skin lesion segmentation. The proposed method is obtained competitive results with the latest skin lesion segmentation methods. The hyperparameters optimized SegNet has achieved the best results with the average Jaccard Index of 84.9 on ISBI2016 and 74.5 on ISBI2017 dataset. Experimental results indicate the validity of Bayesian optimized SegNet. In this study, it has been observed that the bayesian hyperparameter optimization in the SegNet, which is the latest deep learning architecture, increased the segmentation performance of the SegNet by 16% in the ISBI2016 dataset and by 7% in the ISBI2017 dataset.














Similar content being viewed by others
References
Abbas Q, Fondón I, Rashid M (2011) Unsupervised skin lesions border detection via two-dimensional image analysis. Comput Methods Prog Biomed 104:e1–e15. https://doi.org/10.1016/j.cmpb.2010.06.016
Agarwal A, Issac A, Dutta MK, et al (2017) Automated skin lesion segmentation using k-means clustering from digital dermoscopic images. In: 2017 40th international conference on telecommunications and signal processing, TSP 2017. Institute of Electrical and Electronics Engineers Inc., pp 743–748
Ahmed M, B SV (2019) Optimization for facial age estimation. https://doi.org/10.1007/978-3-030-27272-2_21
Ahn E, Bi L, Jung YH, et al (2015) Automated saliency-based lesion segmentation in dermoscopic images. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBS 2015-Novem:3009–3012. https://doi.org/10.1109/EMBC.2015.7319025
Ahn E, Kim J, Bi L, Kumar A, Li C, Fulham M, Feng DD (2017) Saliency-based lesion segmentation via background detection in Dermoscopic images. IEEE J Biomed Heal Informatics 21:1685–1693. https://doi.org/10.1109/JBHI.2017.2653179
Al-masni MA, Al-antari MA, Choi MT et al (2018) Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Prog Biomed 162:221–231. https://doi.org/10.1016/j.cmpb.2018.05.027
Argenziano G, Soyer HP (2001) Dermoscopy of pigmented skin lesions - a valuable tool for early diagnosis of melanoma. Lancet Oncol 2:443–449. https://doi.org/10.1016/S1470-2045(00)00422-8
Argenziano G, Fabbrocini G, Carli P, de Giorgi V, Sammarco E, Delfino M (1998) Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol 134:1563–1570. https://doi.org/10.1001/archderm.134.12.1563
Badrinarayanan V, Kendall A, Cipolla R (2017) SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39:2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615
Barata C, Celebi ME, Marques JS (2015) Improving dermoscopy image classification using color constancy. IEEE J Biomed Heal Informatics 19:1146–1152. https://doi.org/10.1109/JBHI.2014.2336473
Bayesian Optimization Algorithm - MATLAB & Simulink (2020) https://www.mathworks.com/help/stats/bayesian-optimization-algorithm.html. Accessed 20 May 2020
Bi L, Kim J, Ahn E, et al (2016) Automated Skin Lesion Segmentation via Image-wise Supervised Learning and Multi-Scale Superpixel Based Cellular Automata School of Information Technologies, University of Sydney, Australia Sydney Medical School, University of Sydney, Australia Med-X R. 1059–1062
Bi L, Kim J, Ahn E, Kumar A, Fulham M, Feng D (2017) Dermoscopic image segmentation via multistage fully convolutional networks. IEEE Trans Biomed Eng 64:2065–2074. https://doi.org/10.1109/TBME.2017.2712771
Brahmbhatt P, Rajan SN (2019) Skin lesion segmentation using SegNet with binary cross-entropy | papers with code. In: Int. Conf. Artif. Intell. Speech Technol. https://paperswithcode.com/paper/skin-lesion-segmentation-using-segnet-with
Brochu E, Cora VM, de Freitas N (2010) A tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. 1–49
Celebi ME, Kingravi HA, Iyatomi H et al (2008) Border detection in dermoscopy images using statistical region merging. Skin Res Technol 14:347–353. https://doi.org/10.1111/j.1600-0846.2008.00301.x
Dalila F, Zohra A, Reda K, Hocine C (2017) Segmentation and classification of melanoma and benign skin lesions. Optik (Stuttg) 140:749–761. https://doi.org/10.1016/j.ijleo.2017.04.084
Emre Celebi M, Wen Q, Hwang S, Iyatomi H, Schaefer G (2013) Lesion border detection in Dermoscopy images using ensembles of Thresholding methods. Skin Res Technol 19:1–7. https://doi.org/10.1111/j.1600-0846.2012.00636.x
Fan H, Xie F, Li Y, Jiang Z, Liu J (2017) Automatic segmentation of dermoscopy images using saliency combined with Otsu threshold. Comput Biol Med 85:75–85. https://doi.org/10.1016/j.compbiomed.2017.03.025
Finlayson GD, Trezzi E (2004) Shades of gray and colour constancy
Garnavi R, Aldeen M, Celebi ME, Varigos G, Finch S (2011) Border detection in dermoscopy images using hybrid thresholding on optimized color channels. Comput Med Imaging Graph 35:105–115. https://doi.org/10.1016/j.compmedimag.2010.08.001
Gulcu A, Kus Z (2020) Hyper-parameter selection in convolutional neural networks using microcanonical optimization algorithm. IEEE Access 8:52528–52540. https://doi.org/10.1109/ACCESS.2020.2981141
Henning JS, Dusza SW, Wang SQ, Marghoob AA, Rabinovitz HS, Polsky D, Kopf AW (2007) The CASH (color, architecture, symmetry, and homogeneity) algorithm for dermoscopy. J Am Acad Dermatol 56:45–52. https://doi.org/10.1016/j.jaad.2006.09.003
Huang L, Yang YZT (2019) Skin lesion segmentation using object scale-oriented fully convolutional neural networks. Signal, Image Video Process 13:431–438. https://doi.org/10.1007/s11760-018-01410-3
Kasmi R, Mokrani K, Rader RK, Cole JG, Stoecker WV (2016) Biologically inspired skin lesion segmentation using a geodesic active contour technique. Skin Res Technol 22:208–222. https://doi.org/10.1111/srt.12252
Lan G, Tomczak JM, Roijers DM, Eiben AE (2020) Time efficiency in optimization with a bayesian-evolutionary algorithm. arXiv 1:1–13
Lee T, Ng V, Gallagher R, Coldman A, McLean D (1997) Dullrazor®: a software approach to hair removal from images. Comput Biol Med 27:533–543. https://doi.org/10.1016/S0010-4825(97)00020-6
Li Y, Shen L (2018) Skin lesion analysis towards melanoma detection using deep learning network. Sensors (Switzerland) 18:1–16. https://doi.org/10.3390/s18020556
Li X, Aldridge B, Ballerini L, et al (2009) Depth data improves skin lesion segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 5762 LNCS:1100–1107. https://doi.org/10.1007/978-3-642-04271-3_133
Menzies SW, Ingvar C, Crotty KA, McCarthy WH (1996) Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Arch Dermatol 132:1178–1182. https://doi.org/10.1001/archderm.132.10.1178
Ninh QC, Tran TT, Tran TT et al (2019) Skin lesion segmentation based on modification of SegNet neural networks. Proc - 2019 6th NAFOSTED Conf Inf Comput Sci NICS:575–578. https://doi.org/10.1109/NICS48868.2019.9023862
Peng Y, Wang N, Wang Y, Wang M (2019) Segmentation of dermoscopy image using adversarial networks. Multimed Tools Appl 78:10965–10981. https://doi.org/10.1007/s11042-018-6523-2
Shan P, Wang Y, Fu C, Song W, Chen J (2020) Automatic skin lesion segmentation based on FC-DPN. Comput Biol Med 123:103762. https://doi.org/10.1016/j.compbiomed.2020.103762
Snoek J, Larochelle H, Adams RP (2012) Practical Bayesian optimization of machine learning algorithms
Sreena S, Lijiya A (2019) Skin lesion analysis towards melanoma detection. 2019 2nd Int Conf Intell Comput Instrum control Technol ICICICT 2019 32–36. https://doi.org/10.1109/ICICICT46008.2019.8993219
Stolz W, Reimann A, Cognetta AB (1994) ABCD rule of dermatoscopy: a new practical method for early recognition of malignant melanoma
Tang P, Liang Q, Yan X, Xiang S, Sun W, Zhang D, Coppola G (2019) Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. Comput Methods Prog Biomed 178:289–301. https://doi.org/10.1016/j.cmpb.2019.07.005
Tripp MK, Watson M, Balk SJ, Swetter SM, Gershenwald JE (2016) State of the science on prevention and screening to reduce melanoma incidence and mortality: the time is now. CA Cancer J Clin 66:460–480. https://doi.org/10.3322/caac.21352
Valle E, Fornaciali M, Menegola A, Tavares J, Vasques Bittencourt F, Li LT, Avila S (2020) Data, depth, and design: learning reliable models for skin lesion analysis. Neurocomputing 383:303–313. https://doi.org/10.1016/j.neucom.2019.12.003
Xie F, Yang J, Liu J, Jiang Z, Zheng Y, Wang Y (2020) Computer methods and programs in biomedicine skin lesion segmentation using high-resolution convolutional neural network. Comput Methods Prog Biomed 186:105241. https://doi.org/10.1016/j.cmpb.2019.105241
Ye F (2017) Particle swarm optimization-based automatic parameter selection for deep neural networks and its applications in large-scale and high-dimensional data
Yu L, Chen H, Dou Q, Qin J, Heng PA (2017) Automated melanoma recognition in Dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36:994–1004.https://doi.org/10.1109/TMI.2016.2642839
Yuan Y, Chao M, Lo YC (2017) Automatic skin lesion segmentation using deep fully convolutional networks with Jaccard distance. IEEE Trans Med Imaging 36:1876–1886. https://doi.org/10.1109/TMI.2017.2695227
Zalaudek I, Argenziano G, Soyer HP, Corona R, Sera F, Blum A, Braun RP, Cabo H, Ferrara G, Kopf AW, Langford D, Menzies SW, Pellacani G, Peris K, Seidenari S, THE DERMOSCOPY WORKING GROUP (2006) Three-point checklist of dermoscopy: an open internet study. Br J Dermatol 154:431–437. https://doi.org/10.1111/j.1365-2133.2005.06983.x
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This manuscript does not contain any studies with human participants carried out by any of the authors.
Conflict of interest
There is no conflict of interest for authorship.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Şahin, N., Alpaslan, N. & Hanbay, D. Robust optimization of SegNet hyperparameters for skin lesion segmentation. Multimed Tools Appl 81, 36031–36051 (2022). https://doi.org/10.1007/s11042-021-11032-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11032-6