Abstract
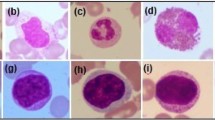
On monitoring an individual's health condition, White Blood Cells play a significant role. The opinion on blood-related disease requires the detection and description of the blood of a patient. Blood cell defects are responsible for numerous health conditions. The conventional technique of manually visualizing White Blood Cells under the microscope is a time-consuming, tedious process and its interpretation requires professionals. There are significant medical applications for an automated method for detecting and classifying blood cells and their subtypes. This work presents an automatic classification method with the help of machine learning for blood cell classification from blood sample medical images. The proposed method can identify and classify the function of each segmented White Blood Cells cell image as granular and non-granular White Blood Cells cell type. It further classifies granular into Eosinophil, Neutrophil and non-granular into Lymphocyte, Monocyte in various forms. Because of its high precision, the proposed framework includes a neural network model to detect white blood cell types. To improve the accuracy of multiple cells overlapping and increase the robustness, data augmentation techniques have been used in the proposed system. Which has improved the accuracy in binary and multi-classification of blood cell subtypes.






Similar content being viewed by others
References
Alam MM, Islam MT (2019) Machine learning approach of automatic identification and counting of blood cells. Healthc Technol Lett 6(4):103–108
Aliyu HA, Sudirman R, Abdul Razak MA, Abd Wahab MA. (2018) Red blood cell classification: deep learning architecture versus support vector machine. In: 2018 2nd international conference on biosignal analysis, processing and systems (ICBAPS), p 142–7
Andrea A, Santiago A, Anna M, Laura P, José R (2019) Recognition of peripheral blood cell images using convolutional neural networks. Comput Methods Programs Biomed 180:105020
Baghel N, Dutta MK, Burget R (2020) Automatic diagnosis of multiple cardiac diseases from PCG signals using convolutional neural network. Comput Methods Programs Biomed 197:105750
Bikhet SF, Darwish AM, Tolba HA, Shaheen SI (2000) Segmentation and classification of white blood cells 2259–61
Burton AG, Jandrey KE (2018) Leukocytosis and Leukopenia. Textbook of Small Animal Emergency Medicine, pp 405–412
Bani-Hani D, Khan N, Alsultan F, Karanjkar S, Nagaru N (2018) Classification of leucocytes using convolutional neural network optimized through genetic algorithm. In: Proceedings of the 7th annual world conference of the Society for Industrial and Systems Engineering, pp 1–7
Daqqa KASA, Maghari AYA, Sarraj WFMA (2017) Prediction and diagnosis of leukemia using classification algorithms. In: 2017 8th international conference on information technology (ICIT), pp 638–43
Gautam A, Singh P, Raman B, Bhadauria H (2016) Automatic classification of leukocytes using morphological features and naive Bayes classifier. In: 2016 IEEE region 10 conference (TENCON), pp 1023–7
Jianwei Z, Minshu Z, Zhenghua Z, Jianjun C, Cao F (2017) Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Comput 55:1287–1301
Kingma D, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint. arXiv:1412.6980
Krishna K, Narasimha MM (1999) Genetic K-means algorithm. IEEE Trans Syst Man Cybern Part B 1999(29):433–439
Kulkarni SS, Hinge CS, Ambekar AG (2013) Classification of RBC and WBC in peripheral blood smear using KNN. Indian J Res
Lawrence S et al (1997) Face recognition: a convolutional neural-network approach. IEEE Trans Neural Netw 1997(8):98–113
Liang G, Hong H, Xie W, Zheng L (2018) Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access 6:36188–36197
Livieris I, Pintelas E, Kanavos A, Pintelas P (2018) Identification of blood cell subtypes from images using an improved SSL algorithm. Biomed J Sci Tech Res 9:6923–6929
Malihi L, Ansari-Asl K, Behbahani A (2013) Malaria parasite detection in giemsastained blood cell images. In: 2013 8th Iranian conference on machine vision and image processing (MVIP), pp 360–5
Medsker LR, Jain LC (2001) Recurrent neural networks. Design Appl 5:64–67
Naz I, Muhammad N, Yasmin M, Sharif M, Shah JH, Fernandes SL (2019) Robust discrimination of leukocytes protuberant types for early diagnosis of leukemia. J Mech Med Biol 19:1950055
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567
Pandey C, Baghel N, Dutta MK, Srivastava A, Choudhary N (2021) Machine learning approach for automatic diagnosis of Chlorosis in Vigna mungo leaves. Multimed Tools Appl 80(9):13407–13427
Patil AM, Patil MD, Birajdar GK (2020) White blood cells image classification using deep learning with canonical correlation analysis. IRBM
Rosyadi T, Arif Nopriadi A, Achmad B, Faridah (2016) Classification of leukocyte images using k-means clustering based on geometry features. In: 2016 6th international annual engineering seminar (InAES), pp 245–9
Safavian SR, David L (1991) (1991) A survey of decision tree classifier methodology. IEEE Trans Syst Man Cybern 21:660–674
Shiqi Y, Jia S, Chunyan X (2017) Convolutional neural networks for hyperspectral image classification. Neurocomputing 219:88–98
Singh A, Khan MA, Baghel N (2020) Face emotion identification by fusing neural network and texture features: facial expression. In: 2020 International Conference on Contemporary Computing and Applications (IC3A), pp 187–190
Sukhia KN, Ghafoor A, Riaz MM, Iltaf N (2019) Automated acute lymphoblastic leukaemia detection system using microscopic images. IET Image Process 13(13):2548–2553
Tai W-L, et al. (2011) Blood cell image classification based on hierarchical SVM. 2011 IEEE International Symposium on Multimedia. IEEE, 2011
Tatdow P, Siripen W, Kittiya K, Aniruth P (2019) Convolutional neural networks for recognition of lymphoblast cell images. Comput Intell Neurosci 2019:1–12
Ushizima DM, Lorena AC, de Carvalho ACPLF (2005) Support vector machines applied to white blood cell recognition. In: Fifth international conference on hybrid intelligent systems (HIS’05), pp 379–84
Young IT (1972) The classification of white blood cells. IEEE Trans Biomed Eng 19(4):291–298
Zahangir Alom Md, Hasan Mahmudul, Yakopcic Chris, Taha Tarek M, Asari Vijayan K (2018) Recurrent residual convolutional neural network based on U-Net (R2UNet) for medical image segmentation. arXiv:1802.06955
Zandecki M et al (2007) Spurious counts and spurious results on haematology analysers: a review. Part II: white blood cells, red blood cells, haemoglobin, red cell indices and reticulocytes. Inter J Lab Hematol 29:21–41
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Baghel, N., Verma, U. & Nagwanshi, K.K. WBCs-Net: type identification of white blood cells using convolutional neural network. Multimed Tools Appl 81, 42131–42147 (2022). https://doi.org/10.1007/s11042-021-11449-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11449-z