Abstract
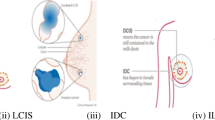
Breast cancer is one of the widespread reasons of morbidity worldwide that begins in the cells of the tissues of morbidity worldwide in the woman community. Breast cancer can be confirmed by investigating the interior tissue regions in terms of Invasive- Ductal-Breast Carcinoma (IDC) and Invasive-lobular-Breast Carcinoma (ILC). Therefore, early diagnosis of breast tissue abnormalities is crucial to diminish the risk by enabling quick and efficient treatment. This research study aims to propose a comprehensive CAD system invasive ductal carcinoma (IDC) by employing the proposed deep learning-based algorithm using the histopathology images. The proposed scheme developed three different CNN models from scratch like ConvNet-A, ConvNet-B, and ConvNet-C by considering different layers that are 8, 9, and 19 layer, respectively. Furthermore, the performance has been validated against four popular machine learning models such as support vector machine (SVM), K-nearest neighbor (KNN), random forest (RF) and logistic regression (LR). Experiments have been performed in two-steps; first, proposed CNN model is evaluated in different sample size, and achieved best accuracy of 88.7% and sensitivity of 92.6% with ConvNet-C with 100,000 sample images. Second, the best classification accuracy achieved by SVM if the number of images taken are more than 5000, because it has a regularization parameter which avoids over-fitting. It is clearly perceived that the proposed CNN algorithms lead to better classification accuracy for detection of IDC compared to state-of-art techniques and hence, this computational framework will act as a helping aid for the pathologist.











Similar content being viewed by others
References
Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N (2016) AggNet: deep learning from crowds for mitosis detection in breast Cancer histology images. IEEE Trans Med Imaging 35(5):1313–1321. https://doi.org/10.1109/TMI.2016.2528120
Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N (2016) AggNet: deep learning from crowds for mitosis detection in breast CancerHistology images. IEEE Trans Med Imaging 35(5):1313–1321. https://doi.org/10.1109/TMI.2016.2528120
Aloyayri A (2020) Breast Cancer Classification from Histopathological Images Using Transfer Learning and Deep Neural Networks School of Graduate Studies. no. April
Bayramoglu N, Heikkilä J (2016) Transfer learning for cell nuclei classification in histopathology images. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 9915 LNCS:532–539. https://doi.org/10.1007/978-3-319-49409-8_46
Bayramoglu N, Heikkilä J (2016) Transfer learning for cell nuclei classificationinhistopathology images. Lect Notes Comput Sci (including SubserLectNotes ArtifIntell Lect Notes Bioinformatics) 9915 LNCS:532–539. https://doi.org/10.1007/978-3-319-49409-8_46
Bayramoglu N, Kannala J, Heikkila J (2016) Deep learning for magnification independent breast cancer histopathology image classification. Proc - Int Conf Pattern Recognit 0:2440–2445. https://doi.org/10.1109/ICPR.2016.7900002
Beevi KS, Nair MS, Bindu GR (2017) A Multi-Classifier System forAutomatic Mitosis Detection in Breast Histopathology Images Using DeepBelief Networks. IEEE J Trans Eng Heal Med 5, no. April. https://doi.org/10.1109/JTEHM.2017.2694004
Conte L, Tafuri B, De Nunzio G, Portaluri M, Galiano A (2020) Breast Cancer Mass detection in DCE-MRI using Deep-Learning Features Followedby Discrimination of Infiltrative vs In Situ Carcinoma through a Machine-learning approach,” no. August. Appl Sci 10:6109. https://doi.org/10.20944/preprints202008.0179.v1
Couture HD et al (2018) Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer 4(1):30. https://doi.org/10.1038/s41523-018-0079-1
Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J (2008) Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features,” 2008 5th IEEE Int. Symp. Biomed. Imaging From Nano to Macro, Proceedings, ISBI, pp. 496–499, https://doi.org/10.1109/ISBI.2008.4541041
Dundar MM, Badve S, Bilgin G, Raykar V, Jain R, Sertel O, Gurcan MN (2011) Computerized classification of intraductal breast lesions using histopathological images. IEEE Trans Biomed Eng 58(7):1977–1984. https://doi.org/10.1109/TBME.2011.2110648
Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, Fan S, Vacek PM, Weaver DL, Herschorn S, Brinton LA, van Ginneken B, Karssemeijer N, Beck AH, Gierach GL, van der Laak JAWM, Sherman ME (2018) Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol 31(10):1502–1512. https://doi.org/10.1038/s41379-018-0073-z
Gecer B, Aksoy S, Mercan E, Shapiro LG, Weaver DL, Elmore JG (2018) Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks. Pattern Recogn 84:345–356. https://doi.org/10.1016/j.patcog.2018.07.022
Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S (2017) Breast Cancer multi-classification from histopathological images with structured deep learning model. Sci Rep 7(1):1–10. https://doi.org/10.1038/s41598-017-04075-z
Kowal M, Filipczuk P, Obuchowicz A, Korbicz J, Monczak R (2013) Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput Biol Med 43(10):1563–1572. https://doi.org/10.1016/j.compbiomed.2013.08.003
Litjens G et al (2016) Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6, no. January:1–11. https://doi.org/10.1038/srep26286
Litjens G et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42, no. December 2012:60–88. https://doi.org/10.1016/j.media.2017.07.005
Loukas C, Kostopoulos S, Tanoglidi A, Glotsos D, Sfikas C, Cavouras D (2013) Breast cancer characterization based on image classification of tissue sections visualized under low magnification. Comput Math Methods Med 2013:1–7. https://doi.org/10.1155/2013/829461
Mobolaji O, Micheal A (2020) Predictive model for likelihood of survival among breast Cancer patients using Machine Learning Techniques. Int J Appl Inf Syst 12(31):29–35
Mohamed ST, Ebied HM, Hassanien A, Tolba E (2019) Optimized Feed Forward Neural Network for Microscopic White Blood Cell. Springer International Publishing
Nguyen CP, Hoang Vo A, Nguyen BT (2019) “Breast Cancer Histology Image classification using Deep Learning,” Proc. - 2019 19th Int. Symp.Commun. Inf. Technol. Isc. 2019, pp. 366–370, https://doi.org/10.1109/ISCIT.2019.8905196
Niwas SI, Palanisamy P, Sujathan K (2010) Wavelet based feature extraction method for Breast cancer cytology images. ISIEA 2010–2010 IEEE Symp. Ind. Electron. Appl., no. Isiea, pp. 686–690, https://doi.org/10.1109/ISIEA.2010.5679377
Osareh A, Shadgar B (2010) Machine learning techniques to diagnose breast cancer. 2010 5th Int. Symp. Heal. Informatics Bioinformatics, HIBIT 2010, pp. 114–120, https://doi.org/10.1109/HIBIT.2010.5478895
Pöllänen I, Braithwaite B, Haataja K, Ikonen T, Toivanen P (2015) Currentanalysis approaches and performance needs for whole slide image processing in breast cancer diagnostics. Proc. - 2015 Int. Conf. Embed. Comput. Syst.Archit. Model. Simulation, SAMOS 2015, no. Samos Xv, pp. 319–325, https://doi.org/10.1109/SAMOS.2015.7363692
Puerto M, Vargas T, Cruz-Roa A (2017) A Digital Pathology application forwhole-slide histopathology image analysis based on genetic algorithm andConvolutional Networks. 2016 IEEE Lat. Am. Conf. Comput.Intell. LA-CCI2016 - Proc., https://doi.org/10.1109/LA-CCI.2016.7885738.
Rachapudi V, Lavanya Devi G (2020) Improved convolutional neural networkbased histopathological image classification. Evol Intell, no. 0123456789 14:1337–1343. https://doi.org/10.1007/s12065-020-00367-y
Sharma S, Mehra R (2020) Conventional machine learning and deep LearningApproach for multi-classification of breast Cancer histopathology images—a comparative insight. J Digit Imaging 33(3):632–654. https://doi.org/10.1007/s10278-019-00307-y
Singh S, Gupta S (2018) Apple scab and Marsonina Coronaria diseases detection in apple leaves using machine learning. Int J Pure Appl Math 118:1151–1166
Sommer C, Fiaschi L, Hamprecht FA, Gerlich DW (2012) Learning-based mitotic cell detection in histopathological images. Proc - Int Conf Pattern Recognit, pp. 2306–2309
Spanhol FA, Oliveira LE, Cavalin PR, Petitjean C, Heutte L (2017) Deep features for breast cancer histopathological image classification. 2017 IEEE international conference on systems, man. Cybern., pp. 1868–1873
Torrents-Barrena J, Puig D, Melendez J, Valls A (2016) Computer-aided diagnosis of breast cancer via Gabor wavelet bank and binary-class SVM in mammographic images. J Exp Theor Artif Intell 28(1–2):295–311. https://doi.org/10.1080/0952813X.2015.1024491
Van Bockstal MR, Berlière M, Duhoux FP, Galant C (2020) Interobserver variability in ductal carcinoma in situ of the breast. Am J Clin Pathol 154:1–14. https://doi.org/10.1093/ajcp/aqaa077
Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, Madabhushi A (2016) Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans Med Imaging 35(1):119–130. https://doi.org/10.1109/TMI.2015.2458702
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not Applicable.
Informed consent
Not Applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, I., Nayak, S.R., Gupta, S. et al. A deep learning based approach to detect IDC in histopathology images. Multimed Tools Appl 81, 36309–36330 (2022). https://doi.org/10.1007/s11042-021-11853-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11853-5