Abstract
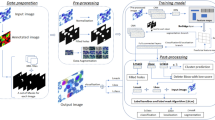
The detection, identification and counting of bone marrow erythroid cells are vital for evaluating the health status and therapeutic schedules of patients with leukemia or hematopathy. However, traditional methods used in hospitals are still based on chemical reagent staining, manual detection and counting with the help of laboratory equipment. And therefore, these methods are time-consuming, laborious, and tedious. The development of deep learning in the field of image processing makes it possible for effective automated detection and classification of erythroid cells. In this research, we proposed a pipeline called ErythroidCounter, which is based on deep learning approaches to perform fully automated detection and classification of erythroid cells. ErythroidCounter is composed of the detection and extraction module (DEM) followed by classification and counting module (CCM). DEM adapts RetinaNet to locate and detect erythroid cells, and it transmits the detected cell images into CCM, while CCM is based on the DenseNet-121 architecture to perform classification and counting., which has close match in terms of classification accuracy compared to manual examination. When classifying erythroid cells, the ErythroidCounter achieved an accuracy of 86.33%, recall of 87.45%, precision of 87.16%, and F1 score of 87.30%. When detecting erythroid cells, ErythroidCounter achieved an precision of 90.7%, recall of 91.3%, and F1 score of 90.9%. EythroidCounter is robust to underlying color images, cell densities, and cell positions. To the best of our knowledge, this is the first automatic approach for erythroid cell detection, classification, and counting in real clinical scenarios, and it can be used as an assistive tool for medical examinations.








Similar content being viewed by others
Change history
08 October 2022
A Correction to this paper has been published: https://doi.org/10.1007/s11042-022-14028-y
References
Acharjee S, Chakrabartty S, Alam MI, Dey N, Santhi V, Ashour AS A semiautomated approach using GUI for the detection of red blood cells. In: 2016 International conference on electrical, electronics, and optimization techniques (ICEEOT), pp 525–529
Acharya V, Kumar P (2018) Identification and red blood cell automated counting from blood smear images using computer-aided system. Med Biol Eng Comput 56.3:483–489
Bhattacharjee R, Saini LM Detection of acute lymphoblastic leukemia using watershed transformation technique. In 2015 International conference on signal processing, computing and control (ISPCC), pp 383–386
Burges CJC (1998) A tutorial on support vector machines for pattern recognition. Data Mining Knowl Discov 2.2:121–167
Choi JW, et al. (2017) White blood cell differential count of maturation stages in bone marrow smear using dual-stage convolutional neural networks. PloS one 12:12
Dai J, Li Y, He K, Sun J (2016) RFCN: object detection via region based fully convolutional networks. In: Conference on neural information processing systems, pp 379–387
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, pp 248–255. IEEE
Erhan D, Szegedy C, Toshev A, Anguelov D (2013) Scalable object detection using deep neural networks. Preprint Available from: arXiv:1312.2249v1
Girshick R (2015) Fast R-CNN. In: Proceedings of the IEEE international conference on computer vision, pp 1440–1448
Girshick R, Donahue J, Darrell T, Malik J (2014) Rich feature hierarchies for accurate object detection and semantic segmentation. Preprint Available from: arXiv:1311.2524v3
Habibzadeh M, Krzyȧk A, Fevens T (2013) White blood cell differential counts using convolutional neural networks for low resolution images. In: International conference on artificial intelligence and soft computing. Springer, Berlin, pp 263–274
Hari J, Sai Prasad A, Koteswara Rao S Separation and counting of blood cells using geometrical features and distance transformed watershed. In: 2014 2nd International conference on devices, circuits and systems (ICDCS), pp 1–5
He K, Zhang X, Ren S, Sun J (2014) Spatial pyramid pooling in deep convolutional networks for visual recognition. In: European conference on computer vision, pp 346–361
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hu J, Shen L, Sun G (2017) Squeeze-and-excitation networks[J]. arXiv:1709.01507
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Jiang M, Cheng L, Qin F, Du L, Zhang M (2018) White blood cells classification with deep convolutional neural networks. Int J Pattern Recognit Artif Intell 32:09
Jiang M, Cheng L, Qin FW, Du L, Zhang M (2018) White blood cells classification with deep convolutional neural networks. Int J Pattern Recogn Artif Intell 32(9):1857006
Kaur P, Sharma V, Garg N Platelet count using image processing. In: 2016 3rd International conference on computing for sustainable global development (INDIACom), pp 2574–2577
Lin TY, Dollár P, Girshick R et al (2017) Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2117–2125
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, pp 2980–2988
Lin T-Y, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, Lawrence Zitnick C (2014) Microsoft coco: common objects in context. In: European conference on computer vision. Springer, Cham, pp 740–755
Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C-Y, Berg AC (2016) SSD: single shot multibox detector. In: European conference on computer vision. Springer, Cham, pp 21–37
Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C, Berg A (2016) SSD: single shot multibox detector. Preprint Available from: arXiv:1512.02325v5
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651
Lou J, Zhou M, Li Q, Yuan C, Liu H An automatic red blood cell counting method based on spectral images. In: 2016 9th International congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI), pp 1391–1396
Qin F, Gao N, Peng Y, Wu Z, Shen S, Grudtsin A (2018) Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput Methods Programs Biomed 162:243–252
Rabbani T, Van Den Heuvel F (2005) Efficient hough transform for automatic detection of cylinders in point clouds. Isprs Wg Iii/3, Iii/4 3:60–65
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real-time object detection. Preprint Available from: arXiv:1506.02640v5
Redmon J, Farhadi A (2018) Yolov3: an incremental improvement. arXiv:1804.02767
Redmon J, Farhadi A (2018) YOLOv3: an incremental improvement. Preprint Available from: arXiv:1804.02767v1
Redmon J, Farhadi A (2016) YOLO9000: better, faster, stronger. Preprint Available from: arXiv:1612.08242v11612.08242v1
Ren S, He K, Girshick R, Sun J, Faster R-CNN (2017) Towards real time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 39(6):1137–1149
Roerdink JBTM, Meijster A (2000) The watershed transform: definitions, algorithms and parallelization strategies. Fundamenta informaticae 41 1 (2):187–228
Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y (2014) OverFeat: integrated recognition, localization and detection using convolutional networks. Preprint Available from: arXiv:1312.6229v4
Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y (2014) OverFeat: integrated recognition, localization and detection using convolutional networks. Preprint Available from: arXiv:1312.6229v4
Shahin AI, et al. (2019) White blood cells identification system based on convolutional deep neural learning networks. Comput Methods Programs Biomedic 168:69–80
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Szegedy C, Toshev A, Erhan D (2013) Deep neural networks for object detection. In: Conference on neural information processing systems, pp 2553–2561
Torrey L, Shavlik J (2010) Transfer learning. In: Handbook of research on machine learning applications and trends: algorithms, methods, and techniques, pp 242–264. IGI global
Venkatalakshmi B, Thilagavathi K Automatic red blood cell counting using hough transform. In: 2013 IEEE Conference on information & communication technologies, pp 267–271
Wang Q, et al. (2019) Deep learning approach to peripheral leukocyte recognition. PloS one 14:6
Xu M, et al. (2017) A deep convolutional neural network for classification of red blood cells in sickle cell anemia. PLoS Comput Biol 13:10
Zhao JW, Zhang MS, Zhou ZH, Chu JJ, Cao FL (2016) Automatic detection and classification of leukocytes using convolutional neural networks. Medical & Biological Engineering & Computing
Zhou M, Wu K, Yu L, et al. (2021) Development and evaluation of a leukemia diagnosis system using deep learning in real clinical scenarios[J]. Front Pediatr 9:693676
Acknowledgements
This research is supported by the National Natural Science Foundation of China (Grants Nos. 61772227, 61972174, 61972175), Science and Technology Development Foundation of Jilin Province (No. 20180201045GX, 20200201300JC, 20200401083GX, 20200201163JC), the Jilin Development and Reform Commission Fund (No. 2020C020-2).
Author information
Authors and Affiliations
Ethics declarations
Conflict of Interests
We declare that the authors have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The corresponding author "You Zhou" in the original publication of this article was incorrect. The corresponding author should be "Liupu Wang".
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, Y., Wang, Y., Wu, J. et al. ErythroidCounter: an automatic pipeline for erythroid cell detection, identification and counting based on deep learning. Multimed Tools Appl 81, 25541–25556 (2022). https://doi.org/10.1007/s11042-022-12209-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12209-3