Abstract
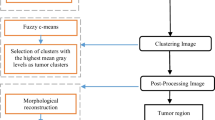
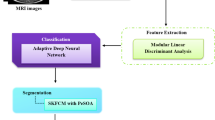
Segmentation of the brain glioma tumour sub-regions is critical to diagnosis and prognosis in clinical applications. This paper proposes a three-stage model to segment automatically brain tumours and their internal tissue in multi-modal brain MRI images. In the first stage, we find the whole tumour (WT) tissue and initially segment it through our neural network model in the FALIR MR sequence. The initial segmentation is fed to the active contour model to segment the precise boundary of WT in the second stage. The combination of the active contour model and the neural network increases the WT segmentation performance significantly. The segmentation of critical internal parts of the tumour, i.e., enhancing tumours (ET), is performed with a fuzzy clustering-based approach in the last stage. Specifically, we apply a multiscale fuzzy C-means (MsFCM) classification method with 12 scales to detect ET in the cropped image of the preceding stage. After segmenting the WT, ET, and normal tissue, we classify input tumours to low-grade glioma (LGG) and high-grade glioma (HGG). For this aim, a local binary pattern algorithm (LBP) and gray-level co-occurrence matrix (GLCM) matrix are used to extract features from the segmented tissues. After creating the feature vectors, we employ the neural network classifier to determine the grade of input tumours in the MR image. The main contributions of the proposed method are: 1) our fuzzy model improved the segmentation accuracy through the designed multiple scales and classes, 2) using tumour edges instead of the entire image as well as utilizing the proposed training algorithm significantly increased the speed of learning and the segmentation accuracy. The experimental results in terms of DICE score indicate that the proposed model is highly competitive compared to state-of-the-art segmentation methods.











Similar content being viewed by others
References
Avital I, Nelkenbaum I, Tsarfaty G, Konen E, Kiryati N, Mayer A (2019) Neural segmentation of seeding ROIs (sROIs) for pre-surgical brain tractography. IEEE Trans Med Imaging 39(5):1655–1667
Chen K, Franko K, Sang R (2021) Structured Model Pruning of Convolutional Networks on Tensor Processing Units. arXiv preprint arXiv:2107.04191
Cho H-h, Park H (2017) Classification of low-grade and high-grade glioma using multi-modal image radiomics features. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp 3081–3084. https://doi.org/10.1109/EMBC.2017.8037508
Chowdhary CL et al (2020) Analytical study of hybrid techniques for image encryption and decryption. Sensors 20(18):5162
Ghahremani M, Ghadiri H, Hamghalam M (2021) Local features integration for content-based image retrieval based on color, texture, and shape. Multimed Tools Appl 80:28245–28263. https://doi.org/10.1007/s11042-021-10895-z
Hamghalam M, Mirzakuchaki S, Ali Akhaee M (2015) Vertex angle image watermarking with optimal detector. Multimed Tools Appl 74:3077–3098. https://doi.org/10.1007/s11042-013-1769-1
Hamghalam M, Wang T, Lei B (2020) High tissue contrast image synthesis via multistage attention-GAN: application to segmenting brain MR scans. Neural Netw 132:43–52. https://doi.org/10.1016/j.neunet.2020.08.014
Hamghalam M, Lei B, Wang T (2020) High tissue contrast MRI synthesis using multi-stage attention-GAN for segmentation. In Proceedings of the AAAI conference on artificial intelligence (Vol. 34, 04, pp. 4067–4074). https://doi.org/10.1609/aaai.v34i04.5825
Hamghalam M, Lei B, Wang T (2020) Convolutional 3D to 2D Patch Conversion for Pixel-Wise Glioma Segmentation in MRI Scans. In: Crimi A, Bakas S (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2019. Lecture notes in computer science, vol 11992. Springer, Cham. https://doi.org/10.1007/978-3-030-46640-4_1
Hamghalam M, Wang T, Qin J, Lei B (2020) Transforming Intensity Distribution of Brain Lesions Via Conditional Gans for Segmentation. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pp 1–4. https://doi.org/10.1109/ISBI45749.2020.9098347
Hamghalam M, Frangi AF, Lei B, Simpson AL (2021) Modality completion via Gaussian process prior Variational autoencoders for multi-modal glioma segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 442–452. https://doi.org/10.1007/978-3-030-87234-2_42
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumour segmentation with deep neural networks. Med Image Anal 35:18–31
Huang M, Yang W, Wu Y, Jiang J, Chen W, Feng Q (2014) Brain tumour segmentation based on local independent projection-based classification. IEEE Trans Biomed Eng 61(10):2633–2645
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1(4):321–331
Ker J, Bai Y, Lee HY, Rao J, Wang L (2019) Automated brain histology classification using machine learning. J Clin Neurosci 66:239–245
Khosravanian A, Rahmanimanesh M, Keshavarzi P, Mozaffari S (2021) Fast level set method for glioma brain tumour segmentation based on Superpixel fuzzy clustering and lattice Boltzmann method. Comput Methods Prog Biomed 198:105809
Lei X, Yu X, Chi J, Wang Y, Zhang J, Wu C (2021) Brain tumour segmentation in MR images using a sparse constrained level set algorithm. Expert Syst Appl 168:114262
Lin WW, Juang C, Yueh MH, Huang TM, Li T, Wang S, Yau ST (2021) 3D brain tumor segmentation using a two-stage optimal mass transport algorithm. Sci Rep 11:14686. https://doi.org/10.1038/s41598-021-94071-1
Lv H, Wang Z, Fu S, Zhang C, Zhai L, Liu X (2017) A robust active contour segmentation based on fractional-order differentiation and fuzzy energy. IEEE Access
Ma C, Luo G, Wang K (2018) Concatenated and connected random forests with multiscale patch driven active contour model for automated brain tumour segmentation of MR images. IEEE Trans Med Imaging 37:1943–1954
Menze BH, Bauer S et al (2015) The multi-modal brain tumour image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024
Mzoughi H, Njeh I, Wali A, Slima MB, BenHamida A, Mhiri C, Mahfoudhe KB (2020) Deep multiscale 3D convolutional neural network (CNN) for MRI gliomas brain tumour classification. J Digit Imaging 33:903–915
Ozkan M, Dawant BM, Maciunas RJ (1993) Neural-network-based segmentation of multi-modal medical images: a comparative and prospective study. IEEE Trans Med Imaging 12(3):534–544
Panigrahi R, Borah S, Bhoi AK, Ijaz MF, Pramanik M, Kumar Y, Jhaveri RH (2021) A consolidated decision tree-based intrusion detection system for binary and multiclass imbalanced datasets. Mathematics. 9(7):751. https://doi.org/10.3390/math9070751
Panigrahi R, Borah S, Bhoi AK, Ijaz MF, Pramanik M, Jhaveri RH, Chowdhary CL (2021) Performance assessment of supervised classifiers for designing intrusion detection systems: a comprehensive review and recommendations for future research. Mathematics. 9(6):690. https://doi.org/10.3390/math9060690
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumour segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251
Polly FP, Shil SK, Hossain MA, Ayman A, Jang YM (2018) Detection and classification of HGG and LGG brain tumor using machine learning. In: 2018 International Conference on Information Networking (ICOIN), pp 813–817. https://doi.org/10.1109/ICOIN.2018.8343231
Soleimany S, Hamghalam M (2017) A novel random-valued impulse noise detector based on MLP neural network classifier. In: 2017 Artificial Intelligence and Robotics (IRANOPEN), pp 165–169. https://doi.org/10.1109/RIOS.2017.7956461
Soleymanifard M, Hamghalam M (2019) Segmentation of Whole Tumor Using Localized Active Contour and Trained Neural Network in Boundaries. In: 2019 5th Conference on Knowledge Based Engineering and Innovation (KBEI), pp 739–744. https://doi.org/10.1109/KBEI.2019.8735050
Srinivasu PN, SivaSai JG, Ijaz MF, Bhoi AK, Kim W, Kang JJ (2021) Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors. 21(8):2852. https://doi.org/10.3390/s21082852
Tamang J et al (2021) Dynamical properties of ion-acoustic waves in space plasma and its application to image encryption - IEEE Access
Wang X-F, Min H, Zou L, Zhang Y-G (2015) A novel level set method for image segmentation by incorporating local statistical analysis and global similarity measurement. Pattern Recogn 48(1):189–204
Zhang K, Zhang L, Lam K-M, Zhang D (2016) A level set approach to image segmentation with intensity inhomogeneity. IEEE Transactions on Cybernetics 46(2):546–557
Zhang D, Huang G, Zhang Q, Han J, Han J, Yu Y (2021) Cross-modality deep feature learning for brain tumour segmentation. Pattern Recogn 110:107562
Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumour segmentation. Medical Image Analysis 43:98–111
Availability of data and material
Our dataset is public and can be downloaded from https://www.smir.ch/BRATS/Start2013
Code availability
Not applicable:
Funding
This work was funded in part by National Institutes of Health R01CA233888.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable:
Consent to participate
Not applicable:
Consent for publication
Not applicable:
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Soleymanifard, M., Hamghalam, M. Multi-stage glioma segmentation for tumour grade classification based on multiscale fuzzy C-means. Multimed Tools Appl 81, 8451–8470 (2022). https://doi.org/10.1007/s11042-022-12326-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12326-z