Abstract
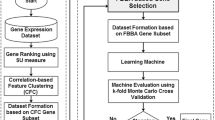
Achieving a satisfactory cancer classification accuracy with the complete set of genes remains a great challenge, due to the high dimensions, small sample size, and presence of noise in gene expression data. Feature reduction is critical and sensitive in the classification task, most importantly in heterogeneous multimedia data. One of the major drawbacks in cancer study is recognizing informative genes from thousands of available genes in microarray data. Traditional feature selection algorithms have failed to scale on large space data like microarray data. Therefore, an effective feature selection algorithm is required to explore the most significant subset of genes by removing non-predictive genes from the dataset without compromising the accuracy of the classification algorithm. The study proposed an information Gain – Modified Bat Algorithm (InfoGain-MBA) features selection model for selecting relevant and informative features from high dimensional Microarray cancer datasets and evaluate the approach with four classifiers - C4.5, Decision Tree, Random Forest and classification and regression tree (CART). The results obtained show that the proposed approach is promising for the classification of microarray cancer data. The random forest has 100% accuracy with few genes in all seven datasets used. Further investigations were also conducted to determine the optimal threshold for each of the datasets.




























Similar content being viewed by others
References
Abeer MM, Basma AM, El-Sayed ME, Abdel-Badeeh MS (2013) Applying a statistical technique for the discovery of differentially expressed genes in microarray data. Int Conf. on Recent Advances in Circuits, Systems, Telecommunications and Control, pp 220–227.
Aitkenhead MJ (2008) A co-evolving decision tree classification method. Expert Syst Appl 34:18–25. https://doi.org/10.1016/j.eswa.2006.08.008
Alomari OA, Khader AT, Al-Betar MA, Abualigah LM (2017) MRMR BA: a hybrid gene selection algorithm for cancer classification. J Theor Appl Inf Technol 95(12):2610–2618
Alshamlan HM, Badr GH, Alohali YA (2015) Genetic bee Colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput Biol Chem 56:49–60. https://doi.org/10.1016/j.compbiolchem.2015.03.001
Bennet J, Ganaprakasam C, Kumar N (2015) A hybrid approach for gene selection and classification using support vector machine. Int Arab J Inf Technol 12(6A):695–700
Biau G (2012) Analysis of a random forests model. J Mach Learn Res 13:1063–1095
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A (2015) Distributed feature selection: an application to microarray data classification. Appl Soft Comput J 30:136–150. https://doi.org/10.1016/j.asoc.2015.01.035
Breiman L, Friedman JH, Olshen RA, Stone CJ (1984) Classification and regression trees, vol 432. Wadsworth International Group, Belmont
Cao J, Zhang L, Wang B, Li F, Yang J (2015) A fast gene selection method for multi-cancer classification using multiple support vector data description. J Biomed Inform 53:381–389
Chormunge S, Jena S (2018) Correlation based feature selection with clustering for high dimensional data. J Electr Syst Inf Technol 5(3):542–549
Chuang LY, Yang CH, Li JC (2011) A hybrid BPSO-CGA approach for gene selection and classification of microarray data. J Comput Biol 19:1–14
Dashtban M, Balafar M, Suravajhala P (2018) Gene selection for tumor classification using a novel bio-inspired multi-objective approach. Genomics 110(1):10–17. https://doi.org/10.1016/j.ygeno.2017.07.010
De Caigny A, Coussement K, De Bock KW (2018) A new hybrid classification algorithm for customer churn prediction based on logistic regression and decision trees. Eur J Oper Res 269(2):760–772. https://doi.org/10.1016/j.ejor.2018.02.009
Dev J, Dash SK, Dash S, Swain M (2012) A classification technique for microarray gene expression data using PSO-FLANN. Int J Comput Sci Eng 4(09):1534–1539
Doddipalli L, Rani KU (2016) Ensemble decision tree classifier for breast Cancer data. Int J Inf Technol Converg Serv 2(1):16–24. https://doi.org/10.5121/ijitcs.2012.2103
Doreswamy H, Salma UM (2016) A binary bat inspired algorithm for the classification of breast Cancer data. Int J Soft Comput Intell Appl 5(2/3):1–21
Ebrahimpour MK, Nezamabadi-Pour H, Eftekhari M (2018) CCFS: a cooperating coevolution technique for large scale feature selection on microarray datasets. Comput Biol Chem 73:171–178
El Akadi A, Amine A, El Ouardighi A, Aboutajdine D (2011) A two-stage gene selection scheme utilizing MRMR filter and GA wrapper. Knowl Inf Syst 26(3):487–500
Fatemeh VS, Sara M, Mohammad HM (2016) A hybrid gene selection approach for microarray data classification using cellular learning automata and ant Colony optimization. Genomics 107:231–238
Forsati R, Moayedikia A, Jensen R, Shamsfard M, Meybodi MR (2014) Enriched ant Colony optimization and its application in feature selection. Neurocomputing 142:354–371
Friedman J, Hastie T, Tibshirani R (2001) The elements of statistical learning, vol 1. Springer Series in Statistics, New York
Gandomi AH, Yang X-S, Alavi AH, Talatahari S (2013) Bat algorithm for constrained optimization tasks. Neural computing and applications. Neural Comput & Applic 22(6):1239–1255
Geetha R, Sivasubramanian S, Kaliappan M, Vimal S, Annamalai S (2019) Cervical cancer identification with synthetic minority oversampling technique and PCA analysis using random forest classifier. J Med Syst 43(9):286
Genuer R, Poggi J-M, Tuleau C (2008) Random Forests: some methodological insights. Retrieved January 18, 2020, from https://doi.org/10.48550/arXiv.0811.3619
Ghorai S, Mukherjee A, Sengupta S, Dutta PK (2010) Cancer classification from gene expression data by NPPC ensemble. IEEE/ACM Trans Comput Biol Bioinforma 8(3):659–671
Griffin DR, Webster FA, Michael CR (1960) The echolocation of flying insects by bats. Animal Behaviour, 8(3):141–154.
Gunavathi C, Premalatha K (2015) Cuckoo search optimisation for feature selection in cancer classification: a new approach. Int J Data Min Bioinform 13(3):248–265
Hall MA, Smith LA (1998) Practical feature subset selection for machine learning. In C. McDonald(Ed.), Computer Science 98 Proceedings of the 21st Australasian Computer Science Conference ACSC’98, Perth, 4-6 February 1998 (pp 181–191). Berlin: Springer.
Hambali MA, Gbolagade MD (2016) Ovarian cancer classification using hybrid synthetic minority over-sampling technique and neural network. J Adv Comput Res 7(4):109–124
Hambali M, Saheed Y, Oladele T, Gbolagade M (2019) ADABOOST ensemble algorithms for breast cancer classification. J Adv Comput Res 10(2):31–52. http://jacr.iausari.ac.ir/article_663924.html. Accessed 18 Jan 2020
Han J, Pei J, Kamber M (2011) Data mining: concepts and techniques. Morgan Kaufmann, 340:94104–3205.
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinforma 2015:1–13. Retrieved January 18, 2020, from https://doi.org/10.1155/2015/198363
Kabir MM, Shahjahan M, Murase K (2012) A new hybrid ant colony optimization algorithm for feature selection. Expert Syst Appl 39(3):3747–3763
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2(3):18–22
Lin S-W, Chen S-C (2012) Parameter determination and feature selection for C4. 5 algorithm using scatter search approach. Soft Comput 16(1):63–75
Lin W-Z, Fang J-A, Xiao X, Chou K-C (2011) iDNA-Prot: identification of DNA binding proteins using random forest with grey model. PLoS One 6(9):e24756
Lønning PE, Sørlie T, Børresen-Dale A-L (2005) Genomics in breast cancer—therapeutic implications. Nat Clin Pract Oncol 2(1):26–33
Mahmoud AM, Maher BA (2014) A hybrid reduction approach for enhancing cancer classification of microarray data. Int J Adv Res Artif Intell 3(10):1–10
Maldonado S, López J (2018) Dealing with high-dimensional class-imbalanced datasets: embedded feature selection for SVM classification. Appl Soft Comput 67:94–105
Martens H (2001) Reliable and relevant modelling of real world data: a personal account of the development of PLS regression. Chemom Intell Lab Syst 58(2):85–95
Martín-Merino M, De Las Rivas J (2009) Improving k-nn for human cancer classification using the gene expression profiles. In: International Symposium on Intelligent Data Analysis, pp. 107–118
Metzner W (1991) Echolocation behaviour in bats. Sci Prog Edinburgh 75(298):453–465. http://www.files/27/ADABOOSTEnsembleAlgorithmsforBreastCancerClassification.ris. Accessed 18 Jan 2020
Mishra S, Shaw K, Mishra D (2012) A new meta-heuristic bat inspired classification approach for microarray data. Procedia Technol 4:802–806
Mitchell TM (1997) Machine learning
Moody J, Darken CJ (1989) Fast learning in networks of locally-tuned processing units. Neural Comput 1(2):281–294
Motieghader H, Najafi A, Sadeghi B, Masoudi-Nejad A (2017) A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Informatics Med Unlocked 9:246–254
Nakamura RYM, Pereira LAM, Costa KA, Rodrigues D, Papa JP, Yang XS (2012) BBA: a binary bat algorithm for feature selection. Braz Symp Comput Graph Image Process 291–297. Retrieved January 18, 2020, from https://doi.org/10.1109/SIBGRAPI.2012.47
Narayanan A, Keedwell EC, Olsson B (2002) Artificial intelligence techniques for bioinformatics. Appl Bioinforma 1:191–222
Nguyen DV, Rocke DM (2002) Tumor classification by partial least squares using microarray gene expression data. Bioinformatics 18(1):39–50
Nguyen DV, Rocke DM (2002) Multi-class cancer classification via partial least squares with gene expression profiles. Bioinformatics 18(9):1216–1226
Panigrahi R, Borah S (2018) Rank allocation to J48 group of decision tree classifiers using binary and multiclass intrusion detection datasets. Procedia Comput Sci 132:323–332
Panigrahi PP, Singh TR (2013) Computational studies on Alzheimer’s disease associated pathways and regulatory patterns using microarray gene expression and network data: revealed association with aging and other diseases. J Theor Biol 334:109–121
Pirooznia M, Yang JY, Yang MQ, Deng Y (2008) A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics 9(S1):S13
Polat K, Güneş S (2009) A novel hybrid intelligent method based on C4. 5 decision tree classifier and one-against-all approach for multi-class classification problems. Expert Syst Appl 36(2):1587–1592
Quinlan JR (1986) Induction of decision trees. Mach Learn 1(1):81–106
Rajeswari P, Reena GS (2011) Human liver cancer classification using microarray gene expression data. Int J Comput Appl 34(6):25–37
Rana MM, Ahmed K (2020) Feature selection and biomedical signal classification using minimum redundancy maximum relevance and artificial neural network. In: Proceedings of International Joint Conference on Computational Intelligence, pp. 207–214
Rangasamy M (2009) An efficient statistical model based classification algorithm for classifying cancer gene expression data with minimal gene subsets. Int J Cyber Soc Educ 2(2):51–66
Revathy N, Amalraj R (2011) Accurate cancer classification using expressions of very few genes. Int J Comput Appl 14(4):19–22
Rodrigues D, Pereira LAM, Nakamura RYM, Costa KAP, Yang XS, Souza AN, Papa JP (2014) A wrapper approach for feature selection based on bat algorithm and optimum-path forest. Expert Syst Appl 41(5):2250–2258
Saeid MM, Nossair ZB, Saleh MA (2020) A microarray cancer classification technique based on discrete wavelet transform for data reduction and genetic algorithm for feature selection. In: 2020 4th International Conference on Trends in Electronics and Informatics (ICOEI)(48184), pp. 857–861
Sahu B, Mishra D (2012) A novel feature selection algorithm using particle swarm optimization for cancer microarray data. Procedia Eng 38:27–31
Schnitzler H-U, Kalko EKV (2001) Echolocation by insect-eating bats: we define four distinct functional groups of bats and find differences in signal structure that correlate with the typical echolocation tasks faced by each group. Bioscience 51(7):557–569
Seera M, Lim CP (2014) A hybrid intelligent system for medical data classification. Expert Syst Appl 41(5):2239–2249
Selvaraj S, Natarajan J (2011) Microarray data analysis and mining tools. Bioinformation 6(3):95
Shafi ASM, Molla MMI, Jui JJ, Rahman MM (2020) Detection of colon cancer based on microarray dataset using machine learning as a feature selection and classification techniques. SN Appl Sci 2(7):1–8
Shreem SS, Abdullah S, Nazri MZA (2014) Hybridising harmony search with a Markov blanket for gene selection problems. Inf Sci (NY) 258:108–121
Sulaiman A, Akinbowale B, Ronke B, Moshood H (2015) Comparative analysis of decision tree algorithms for predicting undergraduate students’ performance in computer programming. J Adv Sci Res Appl 2(20):79–92
Suresh A, Udendhran R, Balamurgan M (2020) Hybridized neural network and decision tree based classifier for prognostic decision making in breast cancers. Soft Comput 24:7947–7953
Swathi S, Babu GA, Sendhilkumar R, Bhukya SN (2012) Performance of ART1 network in the detection of breast cancer. In: Proceedings of International Conference on Computer design and Engineering (ICCDE 2012), vol. 49, pp. 100–105.
Tang R, Fong S, Yang X-S, Deb S (2012) Integrating nature-inspired optimization algorithms to K-means clustering. In: Seventh International Conference on Digital Information Management (ICDIM 2012), pp. 116–123.
Veerabhadrappa, Rangarajan L (2010) Bi-level dimensionality reduction methods using feature selection and feature extraction. Int J Comput Appl 4(2):33–38
Vieira SM, Mendonça LF, Farinha GJ, Sousa JMC (2013) Modified binary PSO for feature selection using SVM applied to mortality prediction of septic patients. Appl Soft Comput 13(8):3494–3504
Wang G, Guo L (2013) A novel hybrid bat algorithm with harmony search for global numerical optimization. J Appl Math vol. 2013. Retrieved January 18, 2020, from https://doi.org/10.1155/2013/696491
Wang L, Chu F, Xie W (2007) Accurate cancer classification using expressions of very few genes. IEEE/ACM Trans Comput Biol Bioinforma 4(1):40–53
Yang X-S (2011) Bat algorithm for multi-objective optimisation. Int J Bio-Inspired Comput 3(5):267–274. https://doi.org/10.1504/IJBIC.2011.042259
Yang X, Gandomi AH (2012) Bat algorithm: a novel approach for global engineering optimization. Eng Comput 29(5):464–483. Retrieved January 18, 2020, from https://doi.org/10.1108/02644401211235834
Yang XS, He X (2013) Bat algorithm: literature review and applications. Int J Bio-Inspired Comput 5(3):141. https://doi.org/10.1504/IJBIC.2013.055093
Zhu Z, Ong Y-S, Dash M (2007) Markov blanket-embedded genetic algorithm for gene selection. Pattern Recogn 40(11):3236–3248
Acknowledgments
This research has been financially supported in part by Tertiary Education Trust Fund (TETFUND) with Reference FUW/REG/T.5/VOL.1/T11. We also acknowledge the support of Ministry of Education of the People’s Republic of China.
Funding
Tertiary Education Trust Fund (TETFUND). Ministry of Education of the People’s Republic of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Not Applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hambali, M.A., Oladele, T.O., Adewole, K.S. et al. Feature selection and computational optimization in high-dimensional microarray cancer datasets via InfoGain-modified bat algorithm. Multimed Tools Appl 81, 36505–36549 (2022). https://doi.org/10.1007/s11042-022-13532-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13532-5