Abstract
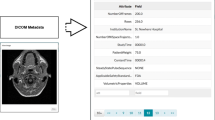
Deep learning has shown record-shattering performance in multiple medical tasks. However, data quantity and quality are crucial requirements. As a matter of fact, data is one of the most challenging issues while deploying deep learning models for different tasks. One of the main challenges is the institutions’ privacy protocols, in particular in the medical field. Indeed, the metadata is usually excluded from the database provided. Many invisible features in images can help tracing anonymized data. We propose to use deep learning to exclude these traces. This article focuses on Magnetic resonance imaging (MRI) and one of the most important features, the equipment used for acquisition. First, we aim to produce an algorithm able to perform well distinguishing multiple MRI equipment from different brands. To this end, we employ a convolution neural network architecture to work on this medical image classification task. The second part of this paper is dedicated to reconstructing the input MRI using a simple auto-encoder. The latter step is to use the auto-encoder in order to mislead the classifier classifying the MRI equipment.











Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Bengio Y (2009) Learning deep architectures for AI. Now Publishers Inc
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828
Bernau D, Grassal P-W, Robl J, Kerschbaum F (2019) Assessing differentially private deep learning with membership inference. arXiv:1912.11328
Chen M, Shi X, Zhang Y, Wu D, Guizani M (2017) Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Transactions on Big Data
Doersch C (2016) Tutorial on variational autoencoders. arXiv:1606.05908
Finlayson S G, Bowers J D, Ito J, Zittrain J L, Beam A L, Kohane I S (2019) Adversarial attacks on medical machine learning. Science 363 (6433):1287–1289
Gatys L A, Ecker A S, Bethge M (2016) Image style transfer using convolutional neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2414–2423
Gillies C E, Taylor D F, Cummings B C, Ansari S, Islim F, Kronick S L, Medlin Jr R P, Ward K R (2020) Demonstrating the consequences of learning missingness patterns in early warning systems for preventative health care: a novel simulation and solution. J Biomed Inform 110:103528
Gondara L (2016) Medical image denoising using convolutional denoising autoencoders. In: 2016 IEEE 16th International conference on data mining workshops (ICDMW). IEEE, pp 241–246
Hamidian S, Sahiner B, Petrick N, Pezeshk A (2017) 3d convolutional neural network for automatic detection of lung nodules in chest ct. In: Medical imaging 2017: computer-aided diagnosis, vol 10134. International Society for Optics and Photonics, p 1013409
Howard J P, Fisher L, Shun-Shin M J, Keene D, Arnold A D, Ahmad Y, Cook C M, Moon J C, Manisty C H, Whinnett Z I et al (2019) Cardiac rhythm device identification using neural networks. JACC: Clin Electrophysiol 5(5):576–586
Jafari H, Omotere O, Adesina D, Wu H-H, Qian L (2018) Iot devices fingerprinting using deep learning. In: MILCOM 2018-2018 IEEE military communications conference (MILCOM). IEEE, pp 1–9
Jeong Y U, Yoo S, Kim Y-H, Shim W H (2020) De-identification of facial features in magnetic resonance images: software development using deep learning technology. J Med Int Res 22(12):e22739
Jordon J, Jarrett D, Yoon J, Elbers P, Thoral P, Ercole A, Zhang C, Belgrave D, van der Schaar M (2020) Hide-and-seek privacy challenge synthetic data generation vs. patient re-identification with clinical time-series data
Kamnitsas K, Ledig C, Newcombe VF, Simpson J P, Kane A D, Menon D K, Rueckert D, Glocker B (2017) Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med Image Anal 36:61–78
Kharrazi M, Sencar H T, Memon N (2004) Blind source camera identification. In: 2004 International conference on image processing, 2004. ICIP’04, vol 1. IEEE, pp 709–712
Kim T, Yang J (2019) Latent-space-level image anonymization with adversarial protector networks. IEEE Access 7:84992–84999. https://doi.org/10.1109/ACCESS.2019.2924479
Kotak J, Elovici Y (2020) Iot device identification using deep learning. In: Conference on complex, intelligent, and software intensive systems. Springer, pp 76–86
Kuppa A, Aouad L, Le-Khac N-A (2021) Towards improving privacy of synthetic datasets. In: Annual privacy forum. Springer, pp 106–119
Lawrence S, Giles C L, Tsoi A C, Back A D (1997) Face recognition: a convolutional neural-network approach. IEEE Trans Neur Netw 8(1):98–113
LeCun Y, Bengio Y, et al. (1995) Convolutional networks for images, speech, and time series. Handbook Brain Theory Neural Netw 3361(10):1995
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Li A, Duan Y, Yang H, Chen Y, Yang J (2020) Tiprdc: task-independent privacy-respecting data crowdsourcing framework for deep learning with anonymized intermediate representations. In: Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining, pp 824–832
Livraga G, Paraboschi S First report on privacy metrics and data sanitisation
Lu X, Tsao Y, Matsuda S, Hori C (2013) Speech enhancement based on deep denoising autoencoder. In: Interspeech, vol 2013, pp 436–440
Makhzani A, Frey B (2013) K-sparse autoencoders. arXiv:1312.5663
Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B (2015) Adversarial autoencoders. arXiv:1511.05644
Merchant K, Revay S, Stantchev G, Nousain B (2018) Deep learning for rf device fingerprinting in cognitive communication networks. IEEE J Selected Topics Signal Process 12(1):160–167
Pereira S, Pinto A, Alves V, Silva C A (2016) Brain tumor segmentation using convolutional neural networks in mri images. IEEE Trans Med Imag 35(5):1240–1251
Price W N, Cohen I G (2019) Privacy in the age of medical big data. Nat Med 25(1):37–43
Riedmiller M, Braun H (1993) A direct adaptive method for faster backpropagation learning: the rprop algorithm. In: IEEE international conference on neural networks. IEEE, pp 586–591
Riyaz S, Sankhe K, Ioannidis S, Chowdhury K (2018) Deep learning convolutional neural networks for radio identification. IEEE Commun Mag 56(9):146–152
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Ryu J, Zheng Y, Gao Y, Abuadbba S, Kim J, Won D, Nepal S, Kim H, Wang C (2021) Can differential privacy practically protect collaborative deep learning inference for the internet of things? arXiv–2104
Shen W, Zhou M, Yang F, Yang C, Tian J (2015) Multi-scale convolutional neural networks for lung nodule classification. In: International conference on information processing in medical imaging. Springer, pp 588–599
Vieira S, Pinaya WH, Mechelli A (2017) Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev 74:58–75
Vinyals O, Blundell C, Lillicrap T, Wierstra D et al (2016) Matching networks for one shot learning. Advances in Neural Information Processing Systems, 29
Wu Q, Feres C, Kuzmenko D, Zhi D, Yu Z, Liu X, et al. (2018) Deep learning based rf fingerprinting for device identification and wireless security. Electron Lett 54(24):1405–1407
Xie L, Lin K, Wang S, Wang F, Zhou J (2018) Differentially private generative adversarial network
Xu R (2020) Functional encryption based approaches for practical privacy-preserving machine learning. Ph.D. Thesis. University of Pittsburgh
Yoon J, Drumright L N, Van Der Schaar M (2020) Anonymization through data synthesis using generative adversarial networks (ads-gan). IEEE J Biomed Health Inform 24(8):2378–2388
Yu D, Zhang H, Chen W, Liu T-Y (2021) Do not let privacy overbill utility: gradient embedding perturbation for private learning. arXiv–2102
Zhao H, Shi J, Qi X, Wang X, Jia J (2017) Pyramid scene parsing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2881–2890
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.;Cogstate;Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.;NeuroRx Research; Neurotrack Technologies;Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics.The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Funding
This study was funded by the University of Poitiers doctoral scholarship.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fezai, L., Urruty, T., Bourdon, P. et al. Deep anonymization of medical imaging. Multimed Tools Appl 82, 9533–9547 (2023). https://doi.org/10.1007/s11042-022-13686-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13686-2