Abstract
Cervical cancer affects more than 500,000 women in the world annually accounting for about 6-9% of all cancer cases, but, its tedious detection procedure makes population-wide screening impossible. Classification of cervical pre-cancerous cells using computer-aided diagnosis tools is a challenging task and is posed an open problem for several decades. The most concerning issue is that only a small amount of data is available publicly. In this study, Deep Learning along with an evolutionary metaheuristic algorithm called the Genetic Algorithm is incorporated for cervical cell classification. Pre-trained Convolutional Neural Networks, namely GoogLeNet and ResNet-18 have been utilized to account for the fewer data available, for extracting deep features from the images. The extracted features are optimized by employing a Genetic Algorithm for feature selection which is coupled with the Support Vector Machines classifier for the final classification. The proposed method has been validated on two publicly available datasets which obtained promising results on 5-fold cross-validation justifying the framework to be reliable. The relevant source codes for the proposed framework has been provided in https://github.com/Rohit-Kundu/Cervical-Cancer-CNN-GAGitHub.










Similar content being viewed by others
References
Alnabelsi SH (2013) Cervical cancer diagnostic system using adaptive fuzzy moving k-means algorithm and fuzzy min-max neural network. J Theor Appl Inform Technol 57:1
Alyafeai Z, Ghouti L (2020) A fully-automated deep learning pipeline for cervical cancer classification. Expert Syst Appl 141:112951
Basak H, Kundu R (2020) Comparative study of maturation profiles of neural cells in different species with the help of computer vision and deep learning. In: International symposium on signal processing and intelligent recognition systems. Springer, pp 352–366
Basak H, Kundu R, Agarwal A, Giri S (2020) Single image super-resolution using residual channel attention network. In: 2020 IEEE 15th International conference on industrial and information systems (ICIIS). IEEE, pp 219–224
Chattopadhyay S, Basak H (2020) Multi-scale attention u-net (msaunet): a modified u-net architecture for scene segmentation. arXiv:200906911
Chattopadhyay S, Dey A, Basak H (2020) Optimizing speech emotion recognition using manta-ray based feature selection. arXiv:200908909
Chattopadhyay S, Dey A, Singh PK, Geem ZW, Sarkar R (2021) Covid-19 detection by optimizing deep residual features with improved clustering-based golden ratio optimizer. Diagnostics 11(2):315
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297
Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, pp 248–255
Dewi EM, Purwanti E, Apsari R (2019) Cervical cell classification using learning vector quantization (lvq) based on shape and statistical features. International Journal of Online and Biomedical Engineering (iJOE) 15 (02):91–98
Dey A, Chattopadhyay S, Singh PK, Ahmadian A, Ferrara M, Sarkar R (2020) A hybrid meta-heuristic feature selection method using golden ratio and equilibrium optimization algorithms for speech emotion recognition. IEEE Access 8:200953–200970
Erlich I, Venayagamoorthy GK, Worawat N (2010) A mean-variance optimization algorithm. In: IEEE Congress on evolutionary computation. IEEE, pp 1–6
Fukushima K, Miyake S (1982) Neocognitron: a self-organizing neural network model for a mechanism of visual pattern recognition. In: Competition and cooperation in neural nets. Springer, pp 267–285
Gandomi AH, Yang XS, Alavi AH (2011) Mixed variable structural optimization using firefly algorithm. Comput Struct 89(23-24):2325–2336
Gill GW (2013) Papanicolaou stain. In: Cytopreparation. Springer, pp 143–189
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Holland JH (1992) Genetic algorithms. Sci Am 267(1):66–73
Hussain E, Mahanta LB, Borah H, Das CR (2020) Liquid based-cytology pap smear dataset for automated multi-class diagnosis of pre-cancerous and cervical cancer lesions. Data in Brief, 105589
Iliyasu AM, Fatichah C (2017) A quantum hybrid pso combined with fuzzy k-nn approach to feature selection and cell classification in cervical cancer detection. Sensors 17(12):2935
Ivakhnenko AG, Lapa VG (1967) Cybernetics and forecasting techniques. North-Holland
Kennedy J, Eberhart R (1995) Particle swarm optimization. In: Proceedings of ICNN’95-international conference on neural networks, vol 4. IEEE, pp 1942–1948
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Li C, Xue D, Zhou X, Zhang J, Zhang H, Yao Y, Kong F, Zhang L, Sun H (2019) Transfer learning based classification of cervical cancer immunohistochemistry images. In: Proceedings of the third international symposium on image computing and digital medicine, pp 102–106
Meyer F (1986) Automatic screening of cytological specimens. Comput Vis Graph Image Process 35(3):356–369
Mirjalili S (2015) Moth-flame optimization algorithm: a novel nature-inspired heuristic paradigm. Knowl-based Syst 89:228–249
Mirjalili S, Lewis A (2016) The whale optimization algorithm. Adv Eng Softw 95:51–67
Mirjalili S, Mirjalili SM, Lewis A (2014) Grey wolf optimizer. Adv Eng Softw 69:46–61
Nguyen N, Poulsen RS, Louis C (1983) Some new color features and their application to cervical cell classification. Pattern Recog 16(4):401–411
Papanicolaou GN, Traut HF (1997) The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Arch Pathol Lab Med 121(3):211
Plissiti ME, Dimitrakopoulos P, Sfikas G, Nikou C, Krikoni O, Charchanti A (2018) Sipakmed: a new dataset for feature and image based classification of normal and pathological cervical cells in pap smear images. In: 2018 25th IEEE International conference on image processing (ICIP). IEEE, pp 3144–3148
Saha R, Bajger M, Lee G (2016) Spatial shape constrained fuzzy c-means (fcm) clustering for nucleus segmentation in pap smear images. In: 2016 international conference on digital image computing: techniques and applications (DICTA). IEEE, pp 1–8
Sevi Ö (2020) Health and Science. Iksad, ISBN. 978-625-7897-22-8
Shi J, Wang R, Zheng Y, Jiang Z, Yu L (2019) Graph convolutional networks for cervical cell classification. Second MICCAI Workshop on Computational Pathology (COMPAT)
Shortliffe EH, Buchanan BG (1975) A model of inexact reasoning in medicine. Math Biosci 23(3-4):351–379
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:14091556
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Tseng CJ, Lu CJ, Chang CC, Chen GD (2014) Application of machine learning to predict the recurrence-proneness for cervical cancer. Neural Comput Applic 24(6):1311–1316
Van Belle V, Pelckmans K, Van Huffel S, Suykens JA (2011) Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif Intell Med 53(2):107–118
William W, Ware A, Basaza-Ejiri AH, Obungoloch J (2018) A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images. Comput Methods Progr Biomed 164:15–22
Win KP, Kitjaidure Y, Hamamoto K, Myo Aung T (2020) Computer-assisted screening for cervical cancer using digital image processing of pap smear images. Appl Sci 10(5):1800
Yang XS, Gandomi AH (2012) Bat algorithm: a novel approach for global engineering optimization. Engineering Computations
Zare MR, Alebiosu DO, Lee SL (2018) Comparison of handcrafted features and deep learning in classification of medical x-ray images. In: 2018 Fourth international conference on information retrieval and knowledge management (CAMP). IEEE, pp 1–5
Zhang L, Kong H, Liu S, Wang T, Chen S, Sonka M (2017) Graph-based segmentation of abnormal nuclei in cervical cytology. Comput Med Imaging Graph 56:38–48
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The article does not contain any studies with human participants performed by any of the authors.
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A: Genetic algorithm
Appendix A: Genetic algorithm
The Genetic Algorithm (GA) is a metaheuristic algorithm inspired by the evolutionary strategy of nature. It incorporates functions like natural selection, crossover and mutation, the factors that are the basic building blocks of evolution. Natural Selection is the procedure by which individuals with the highest fitness, i.e., adaptability with the current state of nature, thrive (“Survival of the fittest” as mentioned by the Father of Evolution, Charles Darwin) and are the ones who create the progeny or off-springs or the next generation. Crossover is the process by which genes from the parents combine to create a new gene, i.e., the offspring. During crossover, some genes mutate or change due to several factors and hence are not identical to the parent’s gene. These same techniques, in a highly abstract version, are used in creating the GA computer code.
GA operates on a population of artificial chromosomes, which are strings in a finite alphabet (binary, for our present work). Each of these chromosomes represents a member of the population in a generation and each such chromosome has a fitness associated with it which represents how good the individual, i.e., the solution is for the current problem.
The GA starts with a randomly generated population of chromosomes each of whose fitness are evaluated. GA incorporates a fitness-based selection of individuals and recombination (crossover) for creating the successor population or individuals of the next generation. During the recombination or crossover, parents are selected and their genetic material combined for producing children’s chromosomes. This process iterates several times and with each successive generation, the average fitness of the population tends to improves until a stopping criterion is met. This procedure makes the GA “evolve” to the best solution for a given problem.
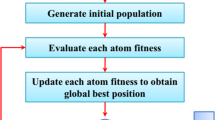
GA is used for the feature selection problem of the present work. Not all the features extracted from the CNN are superior and hence required for the prediction purpose. So, GA is used for selecting the features that are necessary and sufficient to predict the classes of test images and discard the rest. It optimizes the search procedure and takes small computational time. The main components of a GA are chromosome encoding, fitness evaluation, selection of parents, recombination/crossover and the evolution scheme. A more detailed description of these components of GA for classification is presented in Fig. 5.
1.1 A.1 Chromosome encoding
GA manipulates chromosomes of the population in a generation which are string representations of solutions to the problem at hand. This chromosome is an abstraction from the chromosome in biological DNA, which can be considered to be a string of alphabets or numbers. A specific position in the chromosome is called a gene and the character (alphabet or number) that occurs at that location is termed an allele. Any representation used for a given problem is called the GA encoding of that problem (Fig. 11).
The GA used for the present study incorporates a bit-string representation that is the set of possible values of alleles are 0,1. GA is usually applied to a problem where the solution set is finite but, it is so vast that brute-force evaluation is not feasible computationally. In the GA used for the proposed system, the chromosomes are of length the same as the number of features extracted from the CNNs. The bits in the chromosomes represent the selection of a feature, i.e., if a bit (allele) value is 1, it represents that particular feature is selected or “activated”, while a bit value of 0, simply means that feature is not selected or “deactivated”. This is a very simple and straightforward scheme for representation.
1.2 A.2 Initial population
The generation of the initial population is done randomly. The chromosomes are created with positioning bit value 1 at random locations inside the chromosome. Also, to ensure diversity in the population, no individuals with the same chromosome sequence is allowed.
Thus, the initial population has individuals possessing different characteristics since it is guaranteed that different subsets of features will be selected. For the present work, an initial population of size 100 was generated.
1.3 A.3 Fitness of the population
The fitness function is a computation scheme that evaluates the viability of an individual, i.e., a chromosome to be a solution to the given problem. By drawing an analogy with biology, it can be stated that the chromosome sequence is the genotype while the solution it represents is the phenotype. In biological evolution, the chromosomes in a DNA molecule are a set of instructions for the construction of the phenotypical individual/organism. Through a complex series of chemical processes, a small collection of embryonic cells which contain the DNA molecules, are transformed into a fully developed organism, which is then “evaluated” through its success in responding to a variety of environmental factors and influences. Similarly, a GA evaluates the fitness of an individual through a series of scheduled activities that involves a large number of interacting resources. For our present work, the fitness of each individual of the initial population is calculated based on the recognition rate of the SVM classifier on the test samples, before which the classifier is trained with training samples.
1.4 A.4 Selection of parents
GA uses fitness value to discriminate between individuals i.e., the quality of solutions represented by the chromosomes in a generation. The GA’s selection criteria are designed such that the fitness value guides the evolution by selective pressure. So, parents for creating the mating pool is selected based on their fitness values. Selection is usually done with replacement, that is, very fit chromosomes might be selected more than one or even used to recombine with themselves. For the present work, the selection method incorporated is elitism, that is, 50% of the fittest individuals from the current population are selected inside the mating pool for the crossover operation, that is 50 most fit individuals are selected (since the size of the initial population was 100).
1.5 A.5 Crossover or recombination operation
Recombination is the process by which parents’ chromosomes selected from the current population are recombined to form new individuals comprising the new generation. This operation is inspired by the mixing of genetic materials from parents in biological evolution. The idea is that since the parents are selected such that individuals possessing the highest fitness values are inside the mating pool, the recombination of genes from these highly fit parents will hopefully create offspring which have chromosomes with even higher fitness value.
This operation is non-deterministic, it happens with a probability. For our present work, we employ a 1-point crossover. In such a crossover process, two parents are divided into two parts each (usually at the midpoint), and offsprings are created by combining one part from each parent. We used the midpoint crossover technique for the proposed work, an example of which is shown in Fig 10. For our work, the number of off-springs created is calculated by subtracting the number of parents inside the mating pool, from the total number of individuals in the initial population. So, initial population size was 100, and number of parents selected inside mating pool were 50, so the number of offsprings generated = initial population size – number of parents in mating pool = 100 − 50 = 50.
1.6 A.6 Mutation operation
The mutation operator mutates or changes a specific percentage of the total alleles in a generation of chromosomes. This means, a bit is selected at random and if its value was 1 the value is changed to 0 and vice versa, that is a feature selected at random is activated if it was deactivated before and deactivated if it was activated before (Fig. 12).
This operation is also non-deterministic much like the crossover operation and does not guarantee the increase in fitness of a mutated offspring. Mutation rates are usually quite small, and in the present work, a 6% mutation rate was used. An example of the mutation operation is shown in Fig. 13.
1.7 A.7 Successors or next generation
After recombination and mutation, the population of the new generation constitutes the parents that were selected inside the mating pool, that is the 50 fittest individuals of the initial population, and the offsprings created after crossover and mutation operation, that is 50 more individuals. So the new population constitutes 100 individuals (same as the number of individuals in the initial population) whose fitness are again evaluated and this process keeps on repeating until the stopping/termination criterion is met.
1.8 A.8 Termination criterion
The termination criterion for the proposed GA is only the number of generations, i.e., the number of times the whole procedure of population generation, fitness evaluation, offspring generation through crossover and mutation, are repeated. So the tunable parameters for the proposed GA are (i) Size of initial population (ii) Number of parents to be selected inside mating pool (iii) crossover point (iv) Percentage of mutation.
1.9 A.9 Fitness function
The classifiers are embedded in the Genetic Algorithm for fitness assignment to the individuals in the population, i.e., to predict the classes of test images. The accuracy obtained from the classifiers is used as the fitness of the individuals in the population in each generation. Three traditional classifiers have been used and compared in this research which is discussed in the following sections.
Support Vector Machine (SVM)
Support Vector Machine (SVM) is a supervised learning model, which, in a set of training examples, properly labelled with different classes, add new examples to each class making a complete non-probabilistic binary classifier out of this SVM, and is associated with some typical learning algorithms which analyse the data, specifically used for regression and classification tasks. SVM model representation of the training samples in the feature plane is such that a separation between the examples belonging to different classes becomes so prominent, that a line can be fit in that space between two classes which maintain maximum distances from every point of each class and SVM is used to fit that line.
Multi Layer Perceptrons (MLP)
Multi-Layer Perceptron, or MLP is a type of feed-forward Artificial Neural Network (ANN). For MLP, the input is transformed with a non-linear transformation ϕ. The input data is projected to a feature space where it can be linearly separated using this transformation. MLP consists of at least three layers: an input layer, an output layer, and a hidden layer in between input and output layers. MLPs have non-linear activation functions associated with the hidden layers for learning complex patterns in the data. They use gradient descent for forward propagation and utilize back-propagation for network training, which a common supervised learning technique. Having multiple layers and several nodes in each layer allows MLPs to classify even data that are not linearly separable, with high efficiency.
The main drawback of using MLP classifiers is that they have many free parameters in their structure. Usually, these parameters are set using a cross-validation technique which can be fairly time consuming and biased. Therefore, the convergence becomes slower which is not desired.
K-Nearest Neighbours (KNN)
KNN is a non-parametric, supervised Machine Learning algorithm that is used for classification and regression problems. The KNN algorithm stores the feature vectors of the training samples and makes predictions on the test cases based on a user-defined similarity measure, like distance functions. The output class is predicted by a majority voting rule of its K nearest neighbours measured by a distance function.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kundu, R., Chattopadhyay, S. Deep features selection through genetic algorithm for cervical pre-cancerous cell classification. Multimed Tools Appl 82, 13431–13452 (2023). https://doi.org/10.1007/s11042-022-13736-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-13736-9