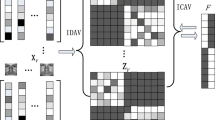
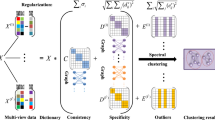
Abstract
Fully capturing valid complementary information in multi-view data enhances the connection between similar data points and weakens the correlation between different data point categories. In this paper, we propose a new multi-view clustering via dual-norm and Hilbert-Schmidt independence criterion (HSIC) induction (MCDHSIC) approach, which can enhance the complementarity, reduce the redundancy between multi-view representations, and improve the accuracy of the clustering results. This model uses the HSIC as the diversity regularization term to capture the nonlinear relationship between different views. In addition, l1-norm and Frobenius norm constraints are imposed to obtain a subspace representation matrix with inter-class sparsity and intra-class consistency. Moreover, we also designed a valuable approach to optimizing the proposed model and theoretically analyzing the convergence of the MCDHSIC method. The results of extensive experiments conducted on five challenging data sets show that the proposed method objectively achieves a highly competent performance compared with several other state-of-the-art multi-view clustering methods.





Similar content being viewed by others
References
Avants BB, Tustison NJ, Stone JR (2021) Similarity-driven multi-view embeddings from high-dimensional biomedical data. Nat Comput Sci 1(2):143–152. https://doi.org/10.1038/s43588-021-00029-8
Bartels RH, Stewart GW (1972) Solution of the matrix equation AX+ XB= C [F4]. Commun ACM 15(9):820–826. https://doi.org/10.1145/361573.361582
Brbić M, Kopriva I (2018) Multi-view low-rank sparse subspace clustering. Pattern Recogn 73:247–258. https://doi.org/10.1016/j.patcog.2017.08.024
Cao X, Zhang C, Fu H et al (2015) Diversity-induced multi-view subspace clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 586–594
Chauhan S, Singh M, Aggarwal AK (2020) An effective health indicator for bearing using corrected conditional entropy through diversity-driven multi-parent evolutionary algorithm. Struct Health Monit 1475921720962419. https://doi.org/10.1177/1475921720962419
Chauhan S, Singh M, Aggarwal AK (2021) Cluster head selection in heterogeneous wireless sensor network using a new evolutionary algorithm. Wirel Pers Commun 119(1):585–616. https://doi.org/10.1007/s11277-021-08225-5
Chauhan S, Singh M, Aggarwal AK (2021) Design of a two-channel quadrature mirror filter bank through a diversity-driven multi-parent evolutionary algorithm. Circuits Syst Signal Process 40(7):3374–3394. https://doi.org/10.1007/s00034-020-01625-1
Chen MS, Huang L, Wang CD, Huang D, Lai JH (2021) Relaxed multi-view clustering in latent embedding space. Inf Fusion 68:8–21. https://doi.org/10.1016/j.inffus.2020.10.013
De Sa VR (2005) Spectral clustering with two views. In: ICML workshop on learning with multiple views, pp. 20–27
Elhamifar E, Vidal R (2013) Sparse subspace clustering: algorithm, theory, and applications. IEEE Trans Pattern Anal Mach Intell 35(11):2765–2781. https://doi.org/10.1109/TPAMI.2013.57
Gretton A, Bousquet O, Smola A, Schölkopf B (2005) Measuring statistical dependence with Hilbert-Schmidt norms. In: International conference on algorithmic learning theory. Springer, Berlin, Heidelberg, pp. 63–77
Hu H, Lin Z, Feng J et al (2014) Smooth representation clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3834–3841
Hu J, Zhou XG, Zhu YH, Yu DJ, Zhang GJ (2019) TargetDBP: accurate DNA-binding protein prediction via sequence-based multi-view feature learning. IEEE/ACM Trans Comput Biol Bioinform 17(4):1419–1429. https://doi.org/10.1109/TCBB.2019.2893634
Hu Z, Nie F, Chang W, Hao S, Wang R et al (2020) Multi-view spectral clustering via sparse graph learning. Neurocomputing 384:1–10. https://doi.org/10.1016/j.neucom.2019.12.004
Huang A, Zhao T, Lin CW (2020) Multi-view data fusion oriented clustering via nuclear norm minimization. IEEE Trans Image Process 29:9600–9613. https://doi.org/10.1109/TIP.2020.3029883
Hussain SF, Khan M, Siddiqi I (2022) Co-clustering based classification of multi-view data. Appl Intell 52:14756–14772. https://doi.org/10.1007/s10489-021-03087-7
Jolliffe IT (2002) Principal component analysis. J Mark Res 87(4):513. https://doi.org/10.2307/3172953
Kumar A, Daumé H (2011) A co-training approach for multi-view spectral clustering. In: Proceedings of the 28th international conference on machine learning, pp 393-400
Lades M, Vorbruggen JC, Buhmann J, Lange J, von der Malsburg C, Wurtz RP, Konen W (1993) Distortion invariant object recognition in the dynamic link architecture. IEEE Trans Comput 42(3):300–311. https://doi.org/10.1109/12.210173
Lee DD, Seung HS (1999) Learning the parts of objects by non-negative matrix factorization. Nature 401(6755):788–791. https://doi.org/10.1038/44565
Li Z, Tang J, He X (2017) Robust structured nonnegative matrix factorization for image representation. IEEE Trans Neural Netw Learn Syst 29(5):1947–1960. https://doi.org/10.1109/TNNLS.2017.2691725
Li Z, Tang J, Mei T (2018) Deep collaborative embedding for social image understanding. IEEE Trans Pattern Anal Mach Intell 41(9):2070–2083. https://doi.org/10.1109/TPAMI.2018.2852750
Lu CY, Min H, Zhao ZQ et al (2012) Robust and efficient subspace segmentation via least squares regression. In: European conference on computer vision. Springer, Berlin, Heidelberg, pp. 347–360
Lu GF, Li H, Wang Y, Tang G (2021) Multi-view subspace clustering with Kronecker-basis-representation-based tensor sparsity measure. Mach Vis Appl 32(6):1–12. https://doi.org/10.1007/s00138-021-01247-w
Ng AY, Jordan MI, Weiss Y (2002) On spectral clustering: analysis and an algorithm. In: Proceedings of the neural information processing systems, pp 849–856
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24(7):971–987. https://doi.org/10.1109/TPAMI.2002.1017623
Pan G, Xiao L, Bai Y, Wilson TW, Stephen JM, Calhoun VD, Wang YP (2020) Multiview diffusion map improves prediction of fluid intelligence with two paradigms of fMRI analysis. IEEE Trans Biomed Eng 68(8):2529–2539. https://doi.org/10.1109/TBME.2020.3048594
Sun Y, Li L, Zheng L, Hu J, Li W, Jiang Y, Yan C (2019) Image classification base on PCA of multi-view deep representation. J Vis Commun Image Represent 62:253–258. https://doi.org/10.1016/j.jvcir.2019.05.016
Turk M, Pentland A (1991) Eigenfaces for recognition. J Cogn Neurosci 3(1):71–86. https://doi.org/10.1162/jocn.1991.3.1.71
Tzortzis G, Likas A (2012) Kernel-based weighted multi-view clustering. In: 2012 IEEE 12th international conference on data mining, pp 675-684
Van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(2605):2579–2605
van Loon W, Fokkema M, Szabo B, de Rooij M (2020) Stacked penalized logistic regression for selecting views in multi-view learning. Inf Fusion 61:113–123. https://doi.org/10.1016/j.inffus.2020.03.007
Wang H, Weng C, Yuan J (2014) Multi-feature spectral clustering with minimax optimization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4106–4113
Wang H, Yang Y, Liu B (2019) GMC: graph-based multi-view clustering. IEEE Trans Knowl Data Eng 32(6):1116–1129. https://doi.org/10.1109/TKDE.2019.2903810
Wang S, Chen Y, Yi S, Chao G (2022) Frobenius norm-regularized robust graph learning for multi-view subspace clustering. Appl Intell 52:14935–14948. https://doi.org/10.1007/s10489-022-03816-6
Wang X, Fu L, Zhang Y, Wang Y et al (2022) MMatch: semi-supervised discriminative representation learning for multi-view classification. IEEE Trans Circuits Syst Video Technol 32:6425–6436. https://doi.org/10.1109/TCSVT.2022.3159371
Xu Y, Wei M (2021) Multi-view clustering toward aerial images by combining spectral analysis and local refinement. Futur Gener Comput Syst 117:138–144. https://doi.org/10.1016/j.future.2020.11.005
Yin Q, Wu S, He R, Wang L (2015) Multi-view clustering via pairwise sparse subspace representation. Neurocomputing 156:12–21. https://doi.org/10.1016/j.neucom.2015.01.017
Zhang C, Fu H, Liu S et al (2015) Low-rank tensor constrained multi-view subspace clustering. In: Proceedings of the IEEE international conference on computer vision, pp. 1582–1590
Zhang C, Hu Q, Fu H et al (2017) Latent multi-view subspace clustering. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4279–4287
Zhang C, Cheng J, Tian Q (2019) Multi-view image classification with visual, semantic and view consistency. IEEE Trans Image Process 29:617–627. https://doi.org/10.1109/TIP.2019.2934576
Acknowledgments
The National Natural Science Foundation of China under Grants 61806006, China Postdoctoral Science Foundation under Grant No. 2019 M660149, the 111 Project under Grants No. B12018, and PAPD of Jiangsu Higher Education Institutions support this work. Guoqing Liu: Conceptualization, Methodology, Validation, Software, Formal analysis, Data curation, Investigation, Writing–original draft. Hongwei Ge: Resources, Review, Supervision, Project administration, Funding acquisition. Shuzhi Su: Review. Shuangxi Wang: Software, Visualization.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, G., Ge, H., Su, S. et al. Multi-view clustering via dual-norm and HSIC. Multimed Tools Appl 83, 36399–36418 (2024). https://doi.org/10.1007/s11042-022-14057-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-14057-7