Abstract
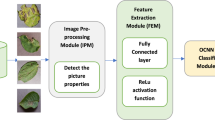
Rice leaf disease, which is a plant disease, causes a decrease in rice production and more importantly, environmental pollution. 10–15% of the losses in rice production are due to rice plant diseases. Automatic recognition of rice leaf disease by computer-assisted expert systems is a promising solution to overcome this problem and to bear the shortage of field experts in this field. Many studies have been conducted using features extracted from deep learning architectures, so far. This study includes keypoint detection on the image, hypercolumn deep feature extraction from CNN layers, and classification stages. The hypercolumn is a vector that contains the activations of all CNN layers for a pixel. Keypoints are prominent points in the images that define what stands out in the image. The first step of the model proposed in this study includes the detection of keypoints on the image and then the extraction of hypercolumn features based on the interest points. In the second step, machine learning experiments are carried out by running classifier algorithms on the features extracted. The evaluation results show that the proposed approach in this paper can detect rice leaf diseases. Furthermore, the Random Forest classifier presented a very successful performance on hypercolumn deep features with 93.06% accuracy, 89.58% sensitivity, 94.79% specificity, and 89.58% precision. As a result, the proposed approach can be integrated into computer-aided rice leaf disease diagnosis systems and so support field experts.








Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Al-Hiary H, Bani-Ahmad S, Reyalat M, et al (2011) Fast and accurate detection and classification of plant diseases
Alshdaifat NFF, Talib AZ, Osman MA (2020) Improved deep learning framework for fish segmentation in underwater videos. Ecol Inform 59:101121. https://doi.org/10.1016/j.ecoinf.2020.101121
Anagnostis A, Tagarakis AC, Asiminari G, Papageorgiou E, Kateris D, Moshou D, Bochtis D (2021) A deep learning approach for anthracnose infected trees classification in walnut orchards. Comput Electron Agric 182:105998. https://doi.org/10.1016/j.compag.2021.105998
Arivazhagan S, Shebiah RN, Ananthi S, Varthini SV (2013) Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features
Bari BS, Islam MN, Rashid M, Hasan MJ, Razman MAM, Musa RM, Ab Nasir AF, Majeed APA (2021) A real-time approach of diagnosing rice leaf disease using deep learning-based faster R-CNN framework. PeerJ Comput Sci 7:e432. https://doi.org/10.7717/PEERJ-CS.432/SUPP-1
Bhattacharya S, Mukherjee A, Phadikar S (2020) A deep learning approach for the classification of Rice leaf diseases. In: Advances in Intelligent Systems and Computing. Springer, pp. 61–69
Capinha C, Ceia-Hasse A, Kramer AM, Meijer C (2021) Deep learning for supervised classification of temporal data in ecology. Ecol Inform:101252. https://doi.org/10.1016/j.ecoinf.2021.101252
Chen J, Zhang D, Zeb A, Nanehkaran YA (2021) Identification of rice plant diseases using lightweight attention networks. Expert Syst Appl 169:114514. https://doi.org/10.1016/j.eswa.2020.114514
Garcia J, Barbedo A (2013) Digital image processing techniques for detecting, quantifying and classifying plant diseases
Ghosal S, Sarkar K (2020) Rice leaf diseases classification using CNN with transfer learning. In: 2020 IEEE Calcutta conference, CALCON 2020 - proceedings. Institute of Electrical and Electronics Engineers Inc., pp 230–236
Gutiérrez S, Hernández I, Ceballos S, Barrio I, Díez-Navajas AM, Tardaguila J (2021) Deep learning for the differentiation of downy mildew and spider mite in grapevine under field conditions. Comput Electron Agric 182:105991. https://doi.org/10.1016/j.compag.2021.105991
Jalal A, Salman A, Mian A, Shortis M, Shafait F (2020) Fish detection and species classification in underwater environments using deep learning with temporal information. Ecol Inform 57:101088. https://doi.org/10.1016/j.ecoinf.2020.101088
Jiang F, Lu Y, Chen Y, Cai D, Li G (2020) Image recognition of four rice leaf diseases based on deep learning and support vector machine. Comput Electron Agric 179:105824. https://doi.org/10.1016/j.compag.2020.105824
Joshi RC, Kaushik M, Dutta MK, Srivastava A, Choudhary N (2021) VirLeafNet: automatic analysis and viral disease diagnosis using deep-learning in Vigna mungo plant. Ecol Inform 61:101197. https://doi.org/10.1016/j.ecoinf.2020.101197
Kahl S, Wood CM, Eibl M, Klinck H (2021) BirdNET: a deep learning solution for avian diversity monitoring. Ecol Inform 61:101236. https://doi.org/10.1016/j.ecoinf.2021.101236
Khirade SD, Patil AB (2015) Plant disease detection using image processing. In: proceedings - 1st international conference on computing, communication, control and automation, ICCUBEA 2015. Institute of Electrical and Electronics Engineers Inc., pp 768–771
Lee SH, Goëau H, Bonnet P, Joly A (2020) New perspectives on plant disease characterization based on deep learning. Comput Electron Agric 170:105220. https://doi.org/10.1016/j.compag.2020.105220
Liu X, Wang C, Bai J, Liao G (2020) Fine-tuning pre-trained convolutional neural networks for gastric precancerous disease classification on magnification narrow-band imaging images. Neurocomputing 392:253–267. https://doi.org/10.1016/J.NEUCOM.2018.10.100
Lu Y, Yi S, Zeng N, Liu Y, Zhang Y (2017) Identification of rice diseases using deep convolutional neural networks. Neurocomputing 267:378–384. https://doi.org/10.1016/J.NEUCOM.2017.06.023
Lumini A, Nanni L (2019) Deep learning and transfer learning features for plankton classification. Ecol Inform 51:33–43. https://doi.org/10.1016/j.ecoinf.2019.02.007
Majeed Y, Zhang J, Zhang X, Fu L, Karkee M, Zhang Q, Whiting MD (2020) Deep learning based segmentation for automated training of apple trees on trellis wires. Comput Electron Agric 170:105277. https://doi.org/10.1016/j.compag.2020.105277
Patidar S, Pandey A, Shirish BA, Sriram A (2020) Rice Plant disease detection and classification using deep residual learning. In: Communications in Computer and Information Science. Springer, pp. 278–293
Peng S, Tang Q, Zou Y (2009) Current status and challenges of rice production in China. Plant Prod Sci 12:3–8
Phadikar S (2012) Classification of Rice leaf diseases based onMorphological changes. International Journal of Information and Electronics Engineering https://doi.org/10.7763/ijiee.2012.v2.137
Prajapati HB, Shah JP, Dabhi VK (2017) Detection and classification of rice plant diseases. Intel Decision Technol 11:357–373. https://doi.org/10.3233/IDT-170301
Ramcharan A, Baranowski K, McCloskey P, Ahmed B, Legg J, Hughes DP (2017) Deep learning for image-based cassava disease detection. Front Plant Sci 8:8. https://doi.org/10.3389/fpls.2017.01852
Rosin PL (1999) Measuring corner properties. Comput Vis Image Underst 73:291–307. https://doi.org/10.1006/CVIU.1998.0719
Rublee E, Rabaud V, Konolige K, Bradski G (2011) ORB: an efficient alternative to SIFT or SURF. In: 2011 international conference on computer vision. IEEE, pp 2564–2571
Sethy PK, Barpanda NK, Rath AK, Behera SK (2020) Deep feature based rice leaf disease identification using support vector machine. Comput Electron Agric 175:105527. https://doi.org/10.1016/j.compag.2020.105527
Sharif H, Hölzel M (2017) A comparison of prefilters in ORB-based object detection. Pattern Recogn Lett 93:154–161. https://doi.org/10.1016/J.PATREC.2016.11.007
Shivali Amit W, Harikrishnan R (2021) A deep learning-based approach in classification and validation of tomato leaf disease. Traitement du signal 38:699–709. https://doi.org/10.18280/TS.380317
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. 3rd international conference on learning representations, ICLR 2015 - conference track proceedings
Tang H, Wang B, Chen X (2020) Deep learning techniques for automatic butterfly segmentation in ecological images. Comput Electron Agric 178:105739. https://doi.org/10.1016/j.compag.2020.105739
Tetila EC, Machado BB, Astolfi G, Belete NAS, Amorim WP, Roel AR, Pistori H (2020) Detection and classification of soybean pests using deep learning with UAV images. Comput Electron Agric 179:105836. https://doi.org/10.1016/j.compag.2020.105836
Tian L, Xue B, Wang Z, Li D, Yao X, Cao Q, Zhu Y, Cao W, Cheng T (2021) Spectroscopic detection of rice leaf blast infection from asymptomatic to mild stages with integrated machine learning and feature selection. Remote Sens Environ 257:112350. https://doi.org/10.1016/j.rse.2021.112350
Toğaçar M, Özkurt KB, Ergen B, Cömert Z (2020) BreastNet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phy A: Statis Mechan App 545:123592. https://doi.org/10.1016/J.PHYSA.2019.123592
Toğaçar M, Özkurt KB, Ergen B, Cömert Z (2020) BreastNet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phy A: Statis Mechan App 545:123592. https://doi.org/10.1016/J.PHYSA.2019.123592
Toğaçar M, Cömert Z, Ergen B (2021) Enhancing of dataset using DeepDream, fuzzy color image enhancement and hypercolumn techniques to detection of the Alzheimer’s disease stages by deep learning model. Neural Comput & Applic 33:1–13. https://doi.org/10.1007/s00521-021-05758-5
Villon S, Mouillot D, Chaumont M, Darling ES, Subsol G, Claverie T, Villéger S (2018) A deep learning method for accurate and fast identification of coral reef fishes in underwater images. Ecol Inform 48:238–244. https://doi.org/10.1016/j.ecoinf.2018.09.007
Xie X, Ma Y, Liu B, He J, Li S, Wang H (2020) A deep-learning-based real-time detector for grape leaf diseases using improved convolutional neural networks. Front Plant Sci 11:751. https://doi.org/10.3389/FPLS.2020.00751/BIBTEX
Xiong Y, Liang L, Wang L, She J, Wu M (2020) Identification of cash crop diseases using automatic image segmentation algorithm and deep learning with expanded dataset. Comput Electron Agric 177:105712. https://doi.org/10.1016/j.compag.2020.105712
Yan Q, Yang B, Wang W, Wang B, Chen P, Zhang J (2020) Apple leaf diseases recognition based on an improved convolutional neural network. Sensors 20:3535. https://doi.org/10.3390/s20123535
Acknowledgements
Author would like to thank Prajapati et al. [25] to provide the public rice leaf dataset.
Sources of funding
No funding is declared.
Declaration of competing interest
The author declares that he has no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akyol, K. Handling hypercolumn deep features in machine learning for rice leaf disease classification. Multimed Tools Appl 82, 19503–19520 (2023). https://doi.org/10.1007/s11042-022-14318-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-14318-5