Abstract
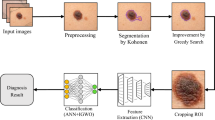
The most dangerous type of skin cancer in the world is Melanoma. Early diagnosis of this cancer in primary stages can increase the chance of surviving death. In recent years, automatic skin cancer detection systems have played a significant role in increasing the rate of cancer diagnosis. Although deep convolutional neural networks presented advantages over traditional methods and brought tremendous breakthroughs in many image classification tasks, accurate classification of skin lesions remains challenging due to the complexity of choosing appropriate architecture for deep neural networks and hyper-parameter tuning. The aim of this paper is to increase the performance of skin lesion classification system through optimizing hyper-parameters and architecture of deep neural network using metaheuristic optimization algorithms. For this purpose, three optimization algorithms are employed to find an optimal configuration for the convolutional neural network either in pre-trained models or model that are trained from scratch. Then the deep features extracted from the optimized models were fused together in pairs and used to train a KNN classifier. The effect of applying hyper-parameter optimization is evaluated on ISIC 2017 and ISIC 2018 datasets. The accuracy of the deep neural network produced by our method reaches to 81.6% and F1-score of 80.9% on ISIC 2017 dataset and accuracy of 90.1% and F1-score of 89.8% on ISIC 2018. The results of the present study indicate that the proposed method outperforms similar methods in classifying seven and three classes of images, without requiring heavy preprocessing and segmentation steps.









Similar content being viewed by others
References
Akram T, Laurent B, Naqvi SR, Alex MM, Muhammad N (2018) A deep heterogeneous feature fusion approach for automatic land-use classification. Inf Sci 467:199–218
Al-Masni MA, Al-Antari MA, Choi M-T, Han S-M, Kim T-S (2018) Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Prog Biomed 162:221–231
Al-Masni MA, Kim D-H, Kim T-S (2020) Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput Methods Prog Biomed 190:105351
Amin J, Sharif A, Gul N, Anjum MA, Nisar MW, Azam F, Bukhari SAC (2020) Integrated design of deep features fusion for localization and classification of skin cancer. Pattern Recogn Lett 131:63–70
Bisla D, Choromanska A, Berman RS, Stein JA, Polsky D (2019) Towards automated melanoma detection with deep learning: Data purification and augmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, pp. 0–0
Bottou L (2010) Large-scale machine learning with stochastic gradient descent. In: Proceedings of COMPSTAT'2010: Springer, pp. 177–186
Chaabouni S, Benois-Pineau J, Amar CB (2016) Transfer learning with deep networks for saliency prediction in natural video. In: 2016 IEEE International Conference on Image Processing (ICIP), IEEE, pp. 1604–1608
Chaturvedi SS, Gupta K, Prasad PS (2020) Skin lesion analyser: An efficient seven-way multi-class skin cancer classification using mobilenet. In: International Conference on Advanced Machine Learning Technologies and Applications, Springer, pp. 165–176
Chen L-C et al (2018) Searching for efficient multi-scale architectures for dense image prediction. Advances in Neural Information Processing Systems, vol. 31
Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1251–1258
Christofides E, Muise A, Desmarais S (2009) Information disclosure and control on Facebook: are they two sides of the same coin or two different processes? Cyberpsychol Behav 12(3):341–345
Chuan L, Quanyuan F (2007) The standard particle swarm optimization algorithm convergence analysis and parameter selection. In: Third International Conference on Natural Computation (ICNC 2007), IEEE, vol. 3, pp. 823–826
Codella NC et al (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), IEEE, pp. 168–172
Cybenko G (1989) Approximation by superpositions of a sigmoidal function. Math Control Signals Syst 2(4):303–314
Dietterich TG (1998) Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput 10(7):1895–1923
Duchi J, Hazan E, Singer Y (2011) Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res 12(7):2121–2159
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. nature 542(7639):115–118
Feurer M, Hutter F (2019) Hyperparameter optimization. In: Automated machine learning. Springer, Cham, pp 3–33
Ghalejoogh GS, Kordy HM, Ebrahimi F (2020) A hierarchical structure based on stacking approach for skin lesion classification. Expert Syst Appl 145:113127
Gibbons JD, Chakraborti S (2014) Nonparametric statistical inference. CRC press
Hameed N, Shabut AM, Ghosh MK, Hossain MA (2020) Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert Syst Appl 141:112961
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778
Hinton G, Srivastava N, Swersky K (2012) Lecture 6a overview of mini–batch gradient descent. Coursera Lecture slides https://class.coursera.org/neuralnets-2012-001/lecture,[Online
Hornik K (1991) Approximation capabilities of multilayer feedforward networks. Neural Netw 4(2):251–257
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708
Hutter F, Kotthoff L, Vanschoren J (2019) Automated machine learning: methods, systems, challenges. Springer Nature
Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, McCullough M, Patel AV, Ma J, Soerjomataram I, Flanders WD, Brawley OW, Gapstur SM, Jemal A (2018) Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin 68(1):31–54
Kennedy J, Eberhart R (1995) Particle swarm optimization. In: Proceedings of ICNN'95-international conference on neural networks, IEEE, vol. 4, pp. 1942–1948
Khan MA, Javed MY, Sharif M, Saba T, Rehman A (2019) Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In: 2019 international conference on computer and information sciences (ICCIS), IEEE, pp. 1–7
Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS (2020) Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recogn Lett 129:293–303
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980
Kittler H, Pehamberger H, Wolff K, Binder M (2002) Diagnostic accuracy of dermoscopy. The lancet oncology 3(3):159–165
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Kulkarni RV, Venayagamoorthy GK (2007) An estimation of distribution improved particle swarm optimization algorithm. In: 2007 3rd international conference on intelligent sensors, sensor networks and information, IEEE, pp. 539–544
Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C (2019) Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph 71:19–29
Marks R (2000) Epidemiology of melanoma: clinical dermatology• review article. Clin Exp Dermatol: Clin Dermatol 25(6):459–463
Masood A, Ali Al-Jumaily A (2013) Computer aided diagnostic support system for skin cancer: a review of techniques and algorithms. Int J Biomed Imaging 2013:1–22
Pan SJ, Yang Q (2009) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Pereira PM et al (2020) Skin lesion classification enhancement using border-line features–the melanoma vs nevus problem. Biomed Signal Process Control 57:101765
Ratul MAR, Mozaffari MH, Lee W-S, Parimbelli E (2020) Skin lesions classification using deep learning based on dilated convolution. BioRxiv, p. 860700
Ribeiro E, Uhl A, Wimmer G, Häfner M (2016) Exploring deep learning and transfer learning for colonic polyp classification. Comput Math Methods Med 2016:1–16
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention, Springer, pp. 234–241
Shi Y, Eberhart RC (1998) Parameter selection in particle swarm optimization. In: International conference on evolutionary programming, Springer, pp. 591–600
Silveira M, Nascimento JC, Marques JS, Marcal AÉRS, Mendonca T, Yamauchi S, Maeda J, Rozeira J (2009) Comparison of segmentation methods for melanoma diagnosis in dermoscopy images. IEEE J Sel Topics Signal Process 3(1):35–45
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Soyer HP, Argenziano G, Zalaudek I, Corona R, Sera F, Talamini R, Barbato F, Baroni A, Cicale L, di Stefani A, Farro P, Rossiello L, Ruocco E, Chimenti S (2004) Three-point checklist of dermoscopy. Dermatology 208(1):27–31
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):1929–1958
Storn R (1995) Differrential evolution-a simple and efficient adaptive scheme for global optimization over continuous spaces, Technical report. International Computer Science Institute, vol. 11
Sun S, Cao Z, Zhu H, Zhao J (2019) A survey of optimization methods from a machine learning perspective. IEEE Trans Cybern 50(8):3668–3681
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2818–2826
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Tan TY, Zhang L, Lim CP (2020) Adaptive melanoma diagnosis using evolving clustering, ensemble and deep neural networks. Knowl-Based Syst 187:104807
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 5(1):1–9
Vanschoren J (2019) Meta-learning. In: Automated Machine Learning. Springer, Cham, pp 35–61
Weiss K, Khoshgoftaar TM, Wang D (2016) A survey of transfer learning. J Big Data 3(1):1–40
Whitley D (1994) A genetic algorithm tutorial. Stat Comput 4(2):65–85
Wistuba M, Schilling N, Schmidt-Thieme L (2016) Hyperparameter optimization machines. In: 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), IEEE, pp. 41–50
Wu G, Lu W, Gao G, Zhao C, Liu J (2016) Regional deep learning model for visual tracking. Neurocomputing 175:310–323
Xie L, Yuille A (2017) Genetic cnn. In: Proceedings of the IEEE international conference on computer vision, pp. 1379–1388
Yan X-H, He F-Z, Chen Y-L (2017) A novel hardware/software partitioning method based on position disturbed particle swarm optimization with invasive weed optimization. J Comput Sci Technol 32(2):340–355
Yang L, Shami A (2020) On hyperparameter optimization of machine learning algorithms: theory and practice. Neurocomputing 415:295–316
Ying C, Klein A, Christiansen E, Real E, Murphy K, Hutter F (2019) Nas-bench-101: Towards reproducible neural architecture search. In: International Conference on Machine Learning, PMLR, pp. 7105–7114
Yu F, Koltun V (2015) Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122
Zeiler MD (2012) ADADELTA: an adaptive learning rate method. CoRR abs/1212.5701 (2012). arXiv preprint arXiv:1212.5701
Zhang N, Cai Y-X, Wang Y-Y, Tian Y-T, Wang X-L, Badami B (2020) Skin cancer diagnosis based on optimized convolutional neural network. Artif Intell Med 102:101756
Zoph B, Le QV (2016) Neural architecture search with reinforcement learning. arXiv preprint arXiv:1611.01578
Zoph B, Vasudevan V, Shlens J, Le QV (2018) Learning transferable architectures for scalable image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8697–8710
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Golnoori, F., Boroujeni, F.Z. & Monadjemi, A. Metaheuristic algorithm based hyper-parameters optimization for skin lesion classification. Multimed Tools Appl 82, 25677–25709 (2023). https://doi.org/10.1007/s11042-023-14429-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14429-7