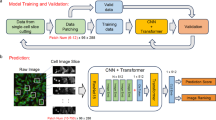
Abstract
Melting curve image is a hallmark of quantitative polymerase chain reaction and is a crucial indicator for the validity of the cycle threshold. Current mainstream methods concentrate on analyzing the melting curve images via artificial process. Therefore, we design a novel multi-scale attention residual network, leveraging various levels space features for accurately classifying the melting curve images. Two modular components are designed in our algorithm. A multi-scale feature extraction module that consists of multi-parallel attention resnet units to selectively capture close related information from various scale feature maps while a series of adaptive multi-scale fusion modules to complete cross-subnet fusion of information. In addition, we also collect massive fluorescence signal data to draw melting curve images for constructing a novel dataset. Our method is evaluated on 3 different benchmark datasets including the self-constructed melting curve image dataset, heartbeat signal dataset and natural color image dataset, a significant highlight is that it achieves a 2.0% accuracy improvement over state-of-the-art in average.






Similar content being viewed by others
Data Availability
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Burger B, Maffettone PM, Gusev VV, Aitchison CM, Bai Y, Wang X, Li X, Alston BM, Li B, Clowes R et al (2020) A mobile robotic chemist. Nature 583:237–241. https://doi.org/10.1038/s41586-020-2442-2
Chakraborty C, Kishor A (2022) Real-time cloud-based patient-centric monitoring using computational health systems. IEEE Trans Comput Social Syst. https://doi.org/10.1109/TCSS.2022.3170375
Chakraborty C, Kishor A, Rodrigues JJ (2022) Novel enhanced-grey wolf optimization hybrid machine learning technique for biomedical data computation. Comput Electr Eng 99:107778. https://doi.org/10.1016/j.compeleceng.2022.107778
Chen X, Cheng Z, Wang S, Lu G, Xv G, Liu Q, Zhu X (2021) Atrial fibrillation detection based on multi-feature extraction and convolutional neural network for processing ecg signals. Comput Methods Programs Biomed 202:106009. https://doi.org/10.1016/j.cmpb.2021.106009
Chen Y, Zhang G, Bai M, Zu S, Guan Z, Zhang M (2019) Automatic waveform classification and arrival picking based on convolutional neural network. Earth Space Sci 6(7):1244–1261. https://doi.org/10.1029/2018EA000466
Coffey KR, Marx RG, Neumaier JF (2019) Deepsqueak: a deep learning-based system for detection and analysis of ultrasonic vocalizations. Neuropsychopharmacology 44:859–868. https://doi.org/10.1038/s41386-018-0303-6
Darmawahyuni A, Nurmaini S, Yuwandini M, Rachmatullah MN, Firdaus F, Tutuko B (2020) Congestive heart failure waveform classification based on short time-step analysis with recurrent network. Inform Med Unlocked 21:100441. https://doi.org/10.1016/j.imu.2020.100441
Dawodi M, Baktash JA, Wada T, Alam N, Joya MZ (2020) Dari speech classification using deep convolutional neural network. In: IEEE International IOT, electronics and mechatronics conference, pp 1–4, DOI https://doi.org/10.1109/IEMTRONICS51293.2020.9216370
Divya B, Subramaniam K, Nanjundaswamy H (2020) Human epithelial type-2 cell image classification using an artificial neural network with hybrid descriptors. IETE J Res 66(1):30–41. https://doi.org/10.1080/03772063.2018.1474810
Duta IC, Liu L, Zhu F, Shao L (2020) Improved residual networks for image and video recognition. In: International conference on pattern recognition, pp 9415–9422
Fki Z, Ammar B, Ayed MB (2021) Optimization of residual convolutional neural network for electrocardiogram classification. arXiv:2112.06024
Fu Q, Xiang L, Zhao K-G, Chen L-Q (2022) Identification and validation of suitable reference genes for quantitative real-time pcr studies in adiantum reniforme var. sinense. J Plant Biochem Biotechnol:1–6. https://doi.org/10.1007/s13562-021-00763-0
Giuste FO, Vizcarra JC (2020) Cifar-10 image classification using feature ensembles. arXiv:2002.03846
Gong Z, Zhong P, Yu Y, Hu W, Li S (2019) A CNN with multiscale convolution and diversified metric for hyperspectral image classification. IEEE Trans Geosci Remote Sens 57(6):3599–3618. https://doi.org/10.1109/TGRS.2018.2886022
Guo Q, Yu X, Ruan G (2019) Lpi radar waveform recognition based on deep convolutional neural network transfer learning. Symmetry 11(4):540. https://doi.org/10.3390/sym11040540
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. European Conf Comput Vision 9908:630–645
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition, pp 770–778
Hou C-Y, Xu T, Zhang L-H, Cui J-T, Zhang Y-H, Li X-S, Zheng L-L, Chen H-Y (2021) Simultaneous detection and differentiation of porcine circovirus 3 and 4 using a sybr green based duplex quantitative pcr assay. J Virol Methods 293:114152. https://doi.org/10.1016/j.jviromet.2021.114152
Howard A, Sandler M, Chu G, Chen L-C, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V et al (2019) Searching for mobilenetv3. In: IEEE international conference on computer vision, pp 1314–1324
Hua W, Zhang C, Xie W, Jin X (2022) Polarimetric SAR image classification based on ensemble dual-branch CNN and superpixel algorithm. IEEE J Sel Top Appl Earth Obs Remote Sens 15:2759–2772. https://doi.org/10.1109/JSTARS.2022.3162953
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: IEEE conference on computer vision and pattern recognition, pp 4700–4708
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) Squeezenet: Alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv:1602.07360
Jia Z, Lin Y, Wang J, Zhou R, Ning X, He Y, Zhao Y (2020) Graphsleepnet: adaptive spatial-temporal graph convolutional networks for sleep stage classification. Int Joint Conference Artif Intell:1324–1330
Kishor A, Chakraborty C (2021) Artificial intelligence and internet of things based healthcare 4.0 monitoring system. Wirel Pers Commun:1–17. https://doi.org/10.1007/s11277-021-08708-5
Kishor A, Chakraborty C, Jeberson W (2021) Reinforcement learning for medical information processing over heterogeneous networks. Multim Tools Appl 80(16):23983–24004. https://doi.org/10.1007/s11042-021-10840-0
Kishor A, Chakraborty C, Jeberson W (2021) Intelligent healthcare data segregation using fog computing with internet of things and machine learning. Int J Eng Syst Model Simul 12(2-3):188–194. https://doi.org/10.1504/IJESMS.2021.115533
Lab S (2021) Heartbeat signal dataset. https://tianchi.aliyun.com/dataset/dataDetail?dataId=94490
Lynn HM, Pan SB, Kim P (2019) A deep bidirectional gru network model for biometric electrocardiogram classification based on recurrent neural networks. IEEE Access 7 145395–145405
Mhathesh T, Andrew J, Sagayam KM, Henesey L (2021) A 3d convolutional neural network for bacterial image classification. Intell Big Data Technol Beyond Hype:419–431. https://doi.org/10.1007/978-981-15-5285-4_42
Müştak İB, Kolukirik M (2021) Development of diagnostic quantitative real-time pcr for mycoplasma gallisepticum and mycoplasma synoviae. Etlik Veteriner Mikrobiyoloji Dergisi 32(1):40–44. https://doi.org/10.35864/evmd.874707
Patel H, Upla KP (2022) A shallow network for hyperspectral image classification using an autoencoder with convolutional neural network. Multim Tools Appl 81(1):695–714. https://doi.org/10.1007/s11042-021-11422-w
Pereira-Gomez M, Fajardo A, Echeverria N, Lopez-Tort F, Perbolianachis P, Costabile A, Aldunate F, Moreno P, Moratorio G (2021) Evaluation of sybr green real time pcr for detecting sars-cov-2 from clinical samples. J Virol Methods 289:114035. https://doi.org/10.1016/j.jviromet.2020.114035
Qiu T, Liu M, Zhou G, Wang L, Gao K (2019) An unsupervised classification method for flame image of pulverized coal combustion based on convolutional auto-encoder and hidden Markov model. Energies 12 (13):2585. https://doi.org/10.3390/en12132585
Ruijter JM, Barnewall RJ, Marsh IB, Szentirmay AN, Quinn JC, van Houdt R, Gunst QD, van den Hoff MJ (2021) Efficiency correction is required for accurate quantitative pcr analysis and reporting. Clin Chem 67(6):829–842. https://doi.org/10.1093/clinchem/hvab052
Sarwinda D, Paradisa RH, Bustamam A, Anggia P (2021) Deep learning in image classification using residual network (resnet) variants for detection of colorectal cancer. Procedia Comput Sci 179:423–431. https://doi.org/10.1016/j.procs.2021.01.025
Shang R, He J, Wang J, Xu K, Jiao L, Stolkin R (2020) Dense connection and depthwise separable convolution based CNN for polarimetric SAR image classification. Knowl Based Syst 194:105542. https://doi.org/10.1016/j.knosys.2020.105542
Sharma M, Bhurane AA, Acharya UR (2022) An expert system for automated classification of phases in cyclic alternating patterns of sleep using optimal wavelet-based entropy features. Expert Syst:12939. https://doi.org/10.1111/exsy.12939
Shi Y, Xu W, Zhang J, Li X (2022) Automated classification of ultrasonic signal via a convolutional neural network. Appl Sci 12(9):4179. https://doi.org/10.3390/app12094179
Simonyan K, Zisserman A (2018) Very deep convolutional networks for large-scale image recognition. Int Conf Learning Representations 75:398–406
Sulistyono MT, Pane ES, Wibawa AD, Purnomo MH (2021) Analysis of eeg-based stroke severity groups clustering using k-means. In: 2021 International seminar on intelligent technology and its applications (ISITIA), pp 67–74. https://doi.org/10.1109/ISITIA52817.2021.9502250. IEEE
Sun C, Chen C, Li W, Fan J, Chen W (2019) A hierarchical neural network for sleep stage classification based on comprehensive feature learning and multi-flow sequence learning. IEEE J Biomed Health Inform 24 (5):1351–1366. https://doi.org/10.1109/JBHI.2019.2937558
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: IEEE conference on computer vision and pattern recognition, pp 1–9
Tan M, Chen B, Pang R, Vasudevan V, Sandler M, Howard A, Le QV (2019) Mnasnet: platform-aware neural architecture search for mobile. In: IEEE conference on computer vision and pattern recognition, pp 2820–2828
Wang X, Huang G, Ma C, Tian W, Gao J (2020) Convolutional neural network applied to specific emitter identification based on pulse waveform images. IET Radar, Sonar Navig 14(5):728–735. https://doi.org/10.1049/iet-rsn.2019.0456
Xiong Z, Nash MP, Cheng E, Fedorov VV, Stiles MK, Zhao J (2018) Ecg signal classification for the detection of cardiac arrhythmias using a convolutional recurrent neural network. Physiol Meas 39(9):094006. https://doi.org/10.1088/1361-6579/aad9ed
Yang M-D, Huang K-H, Tsai H-P (2020) Integrating mnf and hht transformations into artificial neural networks for hyperspectral image classification. Remote Sens 12(14):2327. https://doi.org/10.3390/rs12142327
Yang Y, Hu Y, Zhang X, Wang S (2021) Two-stage selective ensemble of cnn via deep tree training for medical image classification. IEEE Trans Cybern. https://doi.org/10.1109/TCYB.2021.3061147
Yi X-Z, Wu L, Xiang L, Wang M-Y, Chen S-L, Shi Y-H, Liu X (2022) Screening of reference genes for quantitative real-time pcr in artemisia argyi. Zhongguo Zhong yao za zhi= Zhongguo Zhongyao Zazhi= China J Chin Materia Medica 47(3):659–667. https://doi.org/10.19540/j.cnki.cjcmm.20210919.101
Yin B, Corradi F, Bohté S. M. (2021) Accurate and efficient time-domain classification with adaptive spiking recurrent neural networks. Nature Mach Intell 3(10):905–913. https://doi.org/10.1038/s42256-021-00397-w
Yongping L, Xinru Y, Bin W, Mindong C, Jiangting L, Haisheng Z, Qingfang W (2021) Cloning and selection evaluation of reference gene for quantitative real-time pcr in hibiscus esculentus l. J Nucl Agric Sci 35 (1):60. https://doi.org/10.11869/j.issn.100-8551.2021.01.0060
Yu C, Han R, Song M, Liu C, Chang C (2022) Feedback attention-based dense CNN for hyperspectral image classification. IEEE Trans Geosci Remote Sens 60:1–16. https://doi.org/10.1109/TGRS.2021.3058549
Yu H, Sun C, Yang X, Zheng S, Zou H (2019) Fuzzy support vector machine with relative density information for classifying imbalanced data. IEEE Trans Fuzzy Syst 27(12):2353–2367. https://doi.org/10.1109/TFUZZ.2019.2898371
Zhang G, Lin C, Chen Y (2020) Convolutional neural networks for microseismic waveform classification and arrival picking. Geophysics 85(4):227–240. https://doi.org/10.1190/geo2019-0267.1
Zhang J, Wu Y (2017) A new method for automatic sleep stage classification. IEEE Trans Biomed Circ Syst 11(5):1097–1110. https://doi.org/10.1109/TBCAS.2017.2719631
Funding
This research is supported by the National Natural Science Foundation of China (61876070).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, P., Shen, X., Chen, H. et al. M-AResNet: a novel multi-scale attention residual network for melting curve image classification. Multimed Tools Appl 82, 42961–42976 (2023). https://doi.org/10.1007/s11042-023-14694-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14694-6