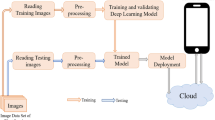
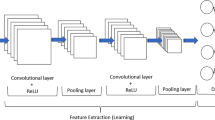
Abstract
Agricultural problems need to be dealt with advanced computing methods to increase food productivity. Automatic classification of plant disease using deep learning methods helps to analyze the food quality and productivity. The existing methods applied Convolutional Neural Network (CNN) based models such as VGG-19, VGG-16, AlexNet, and Resnet-50 for plant disease classification. The existing methods have limitations of vanishing gradient problem and overfitting problem in plant disease classification. The ResNext50-Long Short Term Memory (LSTM) is proposed to improve plant disease classification performance. The hybrid of ResNext50-LSTM model is proposed for effective feature extraction from input images and classification of plant diseases. The ResNext50 architectures helps to limit increases of features in input layer to eliminate bottleneck problem and store relevant features for long term in LSTM for classification. The ResNext 50 model increases the features from 4 to 128 for optimal path construction and existing techniques increases the features more than 128 that causes the overfitting problem. This process of feature limit helps to reduce the overfitting problem and vanishing gradient in LSTM model. The ResNext50 is applied to extract and select the relevant features from input images and LSTM model has advantage of store the relevant information on long term for classification. The ResNext50 model selects the features to differentiate the correlation between the relevant and irrelevant features in feature extraction. The ResNext50 extracted features were applied to the LSTM model to improve classification performance. The proposed Resnext50-LSTM model and existing CNN model performances are tested on plant village dataset to analysis the efficiency. The proposed ResNext50-LSTM model has validation accuracy of 95.44%, and existing ResNext50 model has 93.56% accuracy, and ResNet50 model 93.45% accuracy in plant disease classification.











Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available in the Kaggle repository, https://www.kaggle.com/vipoooool/new-plant-diseases-dataset.
References
Agarwal M, Gupta SK, Biswas KK (2020) Development of efficient CNN model for Tomato crop disease identification. Sustain Comput: Inform Syst 28:100407. https://doi.org/10.1016/j.suscom.2020.100407
Albattah W, Nawaz M, Javed A, Masood M, Albahli S (2022) A novel deep learning method for detection and classification of plant diseases. Complex Intell Syst 8(1):507–524
Barbedo JGA (2018) Impact of dataset size and variety on the effectiveness of deep learning and transfer learning for plant disease classification. Comput Electron Agric 153:46–53. https://doi.org/10.1016/j.compag.2018.08.013
Chen J, Chen J, Zhang D, Sun Y, Nanehkaran YA (2020) Using deep transfer learning for image-based plant disease identification. Comput Electron Agric 173:105393. https://doi.org/10.1016/j.compag.2020.105393
Chen J, Yin H, Zhang D (2020) A self-adaptive classification method for plant disease detection using GMDH-Logistic model. Sustain Comput: Inform Syst 28:100415. https://doi.org/10.1016/j.suscom.2020.100415
Chimmula VKR, Zhang L (2020) Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals 135:109864. https://doi.org/10.1016/j.chaos.2020.109864
Chu Y, Yue X, Yu L, Sergei M, Wang Z (2020) Automatic image captioning based on ResNet50 and LSTM with soft attention. Wirel Commun Mob Comput 2020:8909458. https://doi.org/10.1155/2020/8909458
Ferentinos KP (2018) Deep learning models for plant disease detection and diagnosis. Comput Electron Agric 145:311–318. https://doi.org/10.1016/j.compag.2018.01.009
Gadekallu TR, Rajput DS, Reddy MPK, Lakshmanna K, Bhattacharya S, Singh S, Jolfaei A, Alazab M (2021) A novel PCA–whale optimization-based deep neural network model for classification of tomato plant diseases using GPU. J Real-Time Image Proc 18:1383–1396. https://doi.org/10.1007/s11554-020-00987-8
Hernández S, Lopez JL (2020) Uncertainty quantification for plant disease detection using bayesian deep learning. Appl Soft Comput 96:106597. https://doi.org/10.1016/j.asoc.2020.106597
Jia S, Zhang Y (2018) Saliency-based deep convolutional neural network for no-reference image quality assessment. Multimed Tools Appl 77(12):14859–14872
Kamal KC, Yin Z, Wu M, Wu Z (2019) Depthwise separable convolution architectures for plant disease classification. Comput Electron Agric 165:104948. https://doi.org/10.1016/j.compag.2019.104948
Karthik R, Hariharan M, Anand S, Mathikshara P, Johnson A, Menaka R (2020) Attention embedded residual CNN for disease detection in tomato leaves. Appl Soft Comput 86:105933. https://doi.org/10.1016/j.asoc.2019.105933
Khamparia A, Saini G, Gupta D, Khanna A, Tiwari S, de Albuquerque VHC (2020) Seasonal crops disease prediction and classification using deep convolutional encoder network. Circ Syst Signal Process 39:818–836. https://doi.org/10.1007/s00034-019-01041-0
Khan K, Khan RU, Albattah W, Qamar AM (2022) End-to-end semantic leaf segmentation framework for plants disease classification. Complexity 2022:1168700
Kundu N, Rani G, Dhaka VS, Gupta K, Nayak SC, Verma S, Ijaz MF, Woźniak M (2021) IoT and interpretable machine learning based framework for disease prediction in Pearl Millet. Sensors 21:5386. https://doi.org/10.3390/s21165386
Liu J, Wang X (2020) Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods 16:83. https://doi.org/10.1186/s13007-020-00624-2
Maeda-Gutiérrez V, Galvan-Tejada CE, Zanella-Calzada LA, Celaya-Padilla JM, Galván-Tejada JI, Gamboa-Rosales H, Luna-Garcia H, Magallanes-Quintanar R, Guerrero Mendez CA, Olvera-Olvera CA (2020) Comparison of convolutional neural network architectures for classification of tomato plant diseases. Appl Sci 10:1245. https://doi.org/10.3390/app10041245
Nandhini S, Ashokkumar K (2021) Improved crossover based monarch butterfly optimization for tomato leaf disease classification using convolutional neural network. Multimed Tools Appl 80:18583–18610. https://doi.org/10.1007/s11042-021-10599-4
Saleem MH, Potgieter J, Arif KM (2019) Plant disease detection and classification by deep learning. Plants 8:468. https://doi.org/10.3390/plants8110468
Saleem MH, Khanchi S, Potgieter J, Arif KM (2020) Image-based plant disease identification by deep learning meta-architectures. Plants 9:1451. https://doi.org/10.3390/plants9111451
Saleem MH, Potgieter J, Arif KM (2020) Plant disease classification: a comparative evaluation of convolutional neural networks and deep learning optimizers. Plants 9:1319. https://doi.org/10.3390/plants9101319
Shabbir A, Ali N, Ahmed J, Zafar B, Rasheed A, Sajid M, Ahmed A, Dar SH (2021) Satellite and scene image classification based on transfer learning and fine tuning of resNet50. Math Probl Eng 2021:5843816. https://doi.org/10.1155/2021/5843816
Shah D, Trivedi V, Sheth V, Shah A, Chauhan U (2021) ResTS: residual deep interpretable architecture for plant disease detection. Inform Process Agric. https://doi.org/10.1016/j.inpa.2021.06.001
Shahid F, Zameer A, Muneeb M (2020) Predictions for COVID-19 with deep learning models of LSTM, GRU and Bi-LSTM. Chaos Solitons Fractals 140:110212. https://doi.org/10.1016/j.chaos.2020.110212
Sharma P, Berwal YPS, Ghai W (2020) Performance analysis of deep learning CNN models for disease detection in plants using image segmentation. Inform Process Agric 7:566–574. https://doi.org/10.1016/j.inpa.2019.11.001
Sherstinsky A (2020) Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network. Physica D 404:132306. https://doi.org/10.1016/j.physd.2019.132306
Sujatha R, Chatterjee JM, Jhanjhi NZ, Brohi SN (2021) Performance of deep learning vs machine learning in plant leaf disease detection. Microprocess Microsyst 80:103615. https://doi.org/10.1016/j.micpro.2020.103615
Tian K, Li J, Zeng J, Evans A, Zhang L (2019) Segmentation of tomato leaf images based on adaptive clustering number of K-means algorithm. Comput Electron Agric 165:104962. https://doi.org/10.1016/j.compag.2019.104962
Tran TT, Choi JW, Le TTH, Kim JW (2019) A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant. Appl Sci 9:1601. https://doi.org/10.3390/app9081601
Udutalapally V, Mohanty SP, Pallagani V, Khandelwal V (2021) sCrop: a novel device for sustainable automatic disease prediction, crop selection, and irrigation in internet-of-agro-things for smart agriculture. IEEE Sens J 21:17525–17538. https://doi.org/10.1109/JSEN.2020.3032438
Wang Q, Qi F, Sun M, Qu J, Xue J (2019) Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques. Comput Intell Neurosci 2019:9142753. https://doi.org/10.1155/2019/9142753
Zhang Y, Hutchinson P, Lieven NA, Nunez-Yanez J (2020) Remaining useful life estimation using long short-term memory neural networks and deep fusion. IEEE Access 8:19033–19045
Zhang Y, Song C, Zhang D (2020) Deep learning-based object detection improvement for tomato disease. IEEE Access 8:56607–56614. https://doi.org/10.1109/ACCESS.2020.2982456
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tanwar, S., Singh, J. ResNext50 based convolution neural network-long short term memory model for plant disease classification. Multimed Tools Appl 82, 29527–29545 (2023). https://doi.org/10.1007/s11042-023-14851-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14851-x