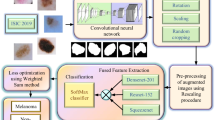
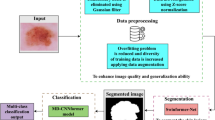
Abstract
Skin cancer is an increasing cause of concern among cancers worldwide. There has been extensive research carried out all over the globe for the early detection of skin cancer to increase the life expectancy of patients. The decision support systems and Computer-aided diagnosis systems aid in detecting cancer at an early stage. The increasing ability of Convolutional Neural Networks (CNN) to extract delicate patterns has made it a popular choice in automated decision support systems. This work proposes a novel U-Net segmentation network with Spatial Attention Blocks (SPAB) called SASegNet to segment the skin lesion accurately. The spatial attention blocks emphasize the model to focus on a particular region. The proposed SASegNet model can provide an accuracy of 95% on the PH2 dataset. In this work, EfficientNet B1 is used for classification. The local features from segmentation results are then passed to EfficientNet B1 to extract features for classification. The pre-processed original images are passed to EfficientNet B1 to extract the global features. Finally, these two features are concatenated to extract the best patterns for classification. Experimentation is carried out on the International Skin Imaging Collaboration (ISIC) datasets. The proposed methodology can obtain the Area Under Curve Receiver Operating Characteristic Curve (AUC-ROC) as 0.974, 0.972, 0.962, and 0.937 for the ISIC-2017, 18, 19, and 2020 datasets. The results obtained are the benchmark results to the best of our knowledge. This automated methodology can aid practising dermatologists in a robust diagnosis.














Similar content being viewed by others
Data availability
All datasets used in the study are publicly available from the ISIC website https://challenge.isic-archive.com/data/#2018.
Abbreviations
- SPAB:
-
SPatial Attention Block
- CNN:
-
Convolutional Neural Networks
- IoU:
-
Intersection over Union
- AUC-ROC:
-
Area Under Curve Receiver Operating Characteristic Curve
- CoE:
-
Center of Excellence
- SPARC:
-
Scheme for Promotion of Academic and Research Collaboration
- MoE:
-
Ministry of Education
- CAD:
-
Computer-Aided Diagnosis
- DL:
-
Deep Learning
- DCNN:
-
Deep Convolutional Neural Networks
- MCSCC:
-
Multi-Class Skin Cancer Classification
- KNN:
-
K- Nearest Neighbors
- SVM:
-
support vector machine
- ANN:
-
artificial neural network
- ISBI:
-
International Symposium on Biomedical Imaging
- ROC:
-
receiver operating characteristic curve
- ISIC:
-
International Skin Imaging Collaboration
- HAM10000:
-
Human Against the Machine
- RCNN:
-
Region-based Convolutional Neural Network
- AUC:
-
area under the curve
- AKIEC:
-
Actinic Keratosis
- BCC:
-
Basal cell carcinoma
- BKL:
-
Benign keratosis
- DF:
-
Dermatofibroma
- NV:
-
Melanocytic nevi
- MEL:
-
Melanoma
- VASC:
-
Vascular lesions
- SCC:
-
Squamous cell carcinoma
- SASegNet:
-
Spatial Attention Segmentation Network
- CM:
-
Confusion Matrix
- TP:
-
True Positive
- TN:
-
True Negative
- FP:
-
False Positive
- FN:
-
False Negative
- DC:
-
Dice Coefficient
- SGD:
-
Stochastic Gradient Descent
- TPR:
-
True Positive Rate
- FPR:
-
False Positive Rate
References
Almseidin M, Abu Zuraiq A, Al-kasassbeh M, Alnidami N (2019) Phishing detection based on machine learning and feature selection methods. Int J Interact Mobile Technol (iJIM) 13(12):171–183. https://doi.org/10.3991/ijim.v13i12.11411
Ballerini L, Fisher RB, Aldridge B, Rees J (2013) A color and texture based hierarchical K-NN approach to the classification of non-melanoma skin lesions. In: Color medical image analysis. Springer, Dordrecht, pp 63–86
Bi L, Kim J, Ahn E, Kumar A, Feng D, Fulham M (2019) Step-wise integration of deep class-specific learning for dermoscopic image segmentation. Pattern Recogn 85:78–89
Cancer Facts and Figures 2021. American Cancer Society. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and statistics/annual-cancer-facts-and-figures/2021/cancer-facts-and-figures-2021.pdf. Accessed January 13, 2021.
Celebi ME, Kingravi HA, Uddin B, Iyatomi H, Aslandogan YA, Stoecker WV, Moss RH (2007) A methodological approach to the classification of dermoscopy images. Comput Med Imaging Graph : Off J Comput Med Imaging Soc 31(6):362–373. https://doi.org/10.1016/j.compmedimag.2007.01.003
Celebi ME, Iyatomi H, Stoecker WV, Moss RH, Rabinovitz HS, Argenziano G, Soyer HP (2008) Automatic detection of blue-white veil and related structures in dermoscopy images. Comput Med Imaging Graphics : Off J Comput Med Imaging Soc 32(8):670–677. https://doi.org/10.1016/j.compmedimag.2008.08.003S
Celebi ME, Iyatomi H, Stoecker WV, Moss RH, Rabinovitz HS, Argenziano G, Soyer HP (2008) Automatic detection of blue-white veil and related structures in dermoscopy images. Comput Med Imaging Graph : Off J Comput Med Imaging Soc 32(8):670–677. https://doi.org/10.1016/j.compmedimag.2008.08.003
Chaturvedi SS, Tembhurne JV, Diwan T (2020) A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl 79(39–40):28477–28498
Chaturvedi SS, Gupta K, Prasad PS (2021) Skin Lesion Analyser: An Efficient Seven-Way Multi-class Skin Cancer Classification Using MobileNet. In: Hassanien A., Bhatnagar R., Darwish A. (eds) Advanced Machine Learning Technologies and Applications. AMLTA 2020. Advances in intelligent systems and computing, vol 1141. Springer, Singapore https://doi.org/10.1007/978-981-15-3383-9_15.
Chen X, Yao L, Zhang Y (2020). Residual Attention U-Net for Automated Multi-Class Segmentation of COVID-19 Chest CT Images. http://arxiv.org/abs/2004.05645
Chen B, Liu Y, Zhang Z, Lu G, Kong AWK (2021) TransAttUNet: Multi-level Attention-guided U-Net with Transformer for Medical Image Segmentation. http://arxiv.org/abs/2107.05274.
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. http://arxiv.org/abs/2102.04306
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, Halpern A (2017) Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). arXiv. https://doi.org/10.48550/arXiv.1710.05006
Combalia M, Codella NC, Rotemberg V, Helba B, Vilaplana V, Reiter O, Carrera C, Barreiro A, Halpern AC, Puig S, Malvehy J (2019) BCN20000: Dermoscopic lesions in the wild. arXiv. https://doi.org/10.48550/arXiv.1908.02288
Dabass M, Vashisth S, Vig R (2021) Attention-Guided deep attrous-residual U-Net architecture for automated gland segmentation in colon histopathology images Informatics in Medicine Unlocked:27. https://doi.org/10.1016/j.imu.2021.100784
Gessert N, … Schlaefer A (2020) Skin lesion classification using CNNs with patch-based attention and diagnosis-guided loss weighting. IEEE Trans Biomed Eng 67(2):495–503
Gessert N, Sentker T, Madesta F, Schmitz R, Kniep H, Baltruschat I, Werner R, Schlaefer A (2020) Skin lesion classification using CNNs with patch-based attention and diagnosis-guided loss weighting. IEEE Trans Biomed Eng 67:495–503
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32. https://doi.org/10.1016/j.jbi.2018.08.006
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32. https://doi.org/10.1016/j.jbi.2018.08.006
Iqbal I, Younus M, Walayat K, Kakar MU, Ma J (2020) Automated multi-class classification of skin lesions through the deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph., vol. 88, no. December, p. 101843
Jain AK, Gupta BB (2016) Comparative analysis of features based machine learning approaches for phishing detection. In: 2016 3rd international conference on computing for sustainable global development (INDIACom), pp 2125–2130
Jha D, Riegler MA, Johansen D, Halvorsen P, Johansen HD (2020) DoubleU-Net: A Deep Convolutional Neural Network for Medical Image Segmentation. http://arxiv.org/abs/2006.04868.
Ji Y, Zhang R, Wang H, Li Z, Wu L, Zhang S, Luo P (2021) Multi-Compound Transformer for Accurate Biomedical Image Segmentation. http://arxiv.org/abs/2106.14385
Jose JM, Sindagi V, Hacihaliloglu I, Patel VM (2020). KiU-Net: Towards Accurate Segmentation of Biomedical Images using Over-complete Representations. http://arxiv.org/abs/2006.04878.
Khan MA, Sharif M, Akram T, Damaševičius R, Maskeliūnas R (2021) Skin Lesion Segmentation and Multiclass Classification Using Deep Learning Features and Improved Moth Flame Optimization. Diagnostics 11, 811(5). https://doi.org/10.3390/diagnostics11050811
Kostopoulos A, Asvestas PA, Kalatzis IK, Sakellaropoulos GC, Sakkis TH, Cavouras DA, Glotsos DT (2017) Adaptable pattern recognition system for discriminating melanocytic nevi from malignant melanomas using plain photography images from different image databases. Int J Med Inform 105:1–10
Krizhevsky A, Sutskever I, Hinton G (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60(6):89–40
Lakhani P, Gray DL, Pett CR, Nagy P, Shih G (2018) Hello world deep learning in medical imaging. J Digit Imaging 31:283–289
Li S, Qin D, Wu X, Li J, Li B, Han W (2022) False alert detection based on deep learning and machine learning. Int J Semantic Web Inform Syst (IJSWIS) 18(1):1–21
Liu L, Mou L, Zhu XX, Mandal M (2020) Automatic skin lesion classification based on mid-level feature learning. Comput Med Imaging Graph:84. https://doi.org/10.1016/j.compmedimag.2020.101765
Liu X, He J, Song L, Liu S, Srivastava G (2021) Medical image classification based on an adaptive size deep learning model. ACM Trans. Multimedia Comput. Commun. Appl. 17, 3s, article 102:18 pages
Maglogiannis I, Doukas CN (2009) Overview of advanced computer vision Systems for Skin Lesions Characterization. IEEE Trans Inform Technol Biomed 13(5):721–733. https://doi.org/10.1109/TITB.2009.2017529
Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C (2019) Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graph 71:19–29. https://doi.org/10.1016/J.COMPMEDIMAG.2018.10.007
Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C (2019) Fusing fine-tuned deep features for skin lesion classification. Comput Med Imaging Graphics : Off J Comput Med Imaging Soc 71:19–29. https://doi.org/10.1016/j.compmedimag.2018.10.007
Mishra A, Gupta BB, Peraković D, Peñalvo FJG, Hsu C-H (2021) Classification Based Machine Learning for Detection of DDoS attack in Cloud Computing. 2021 IEEE Int Conf Consum Electron (ICCE), 1–4 https://doi.org/10.1109/ICCE50685.2021.9427665.
Mubashar M, Ali H, Grönlund C, Azmat S (2022) R2U++: a multiscale recurrent residual U-net with dense skip connections for medical image segmentation. Neural Comput Applic 34:17723–17739. https://doi.org/10.1007/s00521-022-07419-7
Nida N, Irtaza A, Javed A, Yousaf MH, Mahmood MT (2019) Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int J Med Inform 124:37–48. https://doi.org/10.1016/j.ijmedinf.2019.01.005
Nyíri T, Kiss A (2018) Novel Ensembling methods for dermatological image classification. In: International conference on theory and practice of natural computing. Springer, Cham, pp. 438–448.
Oktay O, Schlemper J, le Folgoc L, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D (2018). Attention U-Net: Learning Where to Look for the Pancreas. http://arxiv.org/abs/1804.03999.
Punn NS, Agarwal S (2022) RCA-IUNet: a residual cross-spatial attention-guided inception U-net model for tumor segmentation in breast ultrasound imaging. Mach Vis Appl 33(2). https://doi.org/10.1007/s00138-022-01280-3
Qin X, Zhang Z, Huang C, Dehghan M, Zaiane OR, Jagersand M (2020) U2-net: going deeper with nested U-structure for salient object detection. https://doi.org/10.1016/j.patcog.2020.107404
Rahman Z, Hossain MS, Islam MR, Hasan MM, Hridhee RA (2021) An approach for multiclass skin lesion classification based on ensemble learning. Informatics Med Unlocked 25:100659
Ramella G (2021) Saliency-based segmentation of dermoscopic images using colour information, computer methods in biomechanics and biomedical. Engineering, Imaging & Visualization
Ratul AR, Mozaffari MH, Lee WS, Parimbelli E (2019) Skin lesions classification using deep learning based on dilated convolution bioRxiv:860700. https://doi.org/10.1101/860700
Rebouças Filho PP, Peixoto SA (2018) Automatic histologically-closer classification of skin lesions. Comput. Med. Imaging Graph 68:40–54. https://doi.org/10.1016/j.compmedimag.2018.05.004
Rehman A, Khan MA, Mehmood Z, Saba T, Sardaraz M, Rashid M (2020 Apr) Microscopic melanoma detection and classification: a framework of pixel-based fusion and multilevel features reduction. Microsc Res Tech 83(4):410–423. https://doi.org/10.1002/jemt.23429
Saba T, Khan MA, Rehman A, Marie-Sainte SL (2019) Region extraction and classification of skin cancer: A heterogeneous framework of deep CNN features fusion and reduction. J Med Syst:43
Shahin AH, Kamal, A, Elattar, MA (2018) Deep ensemble learning for skin lesion classification from dermoscopic images. In: IEEE 9th Cairo international biomedical engineering conference - CIBEC’2018, pp 150–153. https://doi.org/10.1109/CIBEC.2018.8641815.
Shahin AH, Kamal A, Elattar MA (2018) Deep ensemble learning for skin lesion classification from dermoscopic images. In: IEEE 9th Cairo international biomedical engineering conference - CIBEC’2018, pp 150–153. https://doi.org/10.1109/CIBEC.2018.8641815.
Song L, Liu X, Chen S, Liu S, Liu X, Muhammad K, Bhattacharyya S (2022) A deep fuzzy model for diagnosis of COVID-19 from CT images. Appl Soft Comput 122:108883. https://doi.org/10.1016/j.asoc.2022.108883
Song L, Liu X, Chen S, Liu S, Liu X, Muhammad K, Bhattacharyya S (2022) A deep fuzzy model for diagnosis of COVID-19 from CT images. Appl Soft Comput 122:108883. https://doi.org/10.1016/j.asoc.2022.108883
Stergiou CL, Plageras AP, Psannis KE, Gupta BB (2020) Secure machine learning scenario from big data in cloud computing via internet of things network. In: Gupta B, Perez G, Agrawal D, Gupta D (eds) Handbook of computer networks and cyber security. Springer, Cham. https://doi.org/10.1007/978-3-030-22277-2_21
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021 May) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Tan M, Le QV (2019) EfficientNet: rethinking model scaling for convolutional neural networks. arXiv. https://doi.org/10.48550/arXiv.1905.11946
Tomar NK, Jha D, Riegler MA, Johansen HD, Johansen D, Rittscher J, Halvorsen P, Ali S (2021). FANet: A Feedback Attention Network for Improved Biomedical Image Segmentation. http://arxiv.org/abs/2103.17235.
Tschandl P, Rosendahl C, Kittler H (2018) The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data 5:180161. https://doi.org/10.1038/sdata.2018.161
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: convolutional block attention module. In proceedings of the European conference on computer vision (ECCV) (pp. 3-19).
Xie Y, Zhang J, Xia Y, Shen C (2020) A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans Med Imaging 39(7):2482–2493
Yuan Y, Lo YC (2019) Improving Dermoscopic image segmentation with enhanced convolutional-Deconvolutional networks. IEEE J Biomed Heal Informatics 23(2):519–526
Zagoruyko S, Komodakis N (2016) Paying more attention to attention: improving the performance of convolutional neural networks via attention transfer. arXiv. https://doi.org/10.48550/arXiv.1612.03928.
Zhao P, Zhang J, Fang W, Deng S (2020) SCAU-net: Spatial-Channel attention U-net for gland segmentation. Frontiers in Bioengineering and Biotechnology 8. https://doi.org/10.3389/fbioe.2020.00670
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) UNet++: a nested u-net architecture for medical image segmentation. Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics). LNCS 11045:3–11. https://doi.org/10.1007/978-3-030-00889-5_1
Acknowledgements
The authors would like to thank the Center of Excellence (CoE) for the Artificial Intelligence Lab at the National Institute of Technology Tiruchirappalli, Tamil Nadu, India, for providing the computational resources.
Funding
This research work was partly funded by the Scheme for Promotion of Academic and Research Collaboration (SPARC), Ministry of Education (MoE) Government of India under grant id SPARC-P641/2019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix A
Appendix A
EfficientNet B1 architecture. In Fig. 18, ×2 and ×3 indicates the blocks are repeated twice and thrice, respectively
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kadirappa, R., S., D., R., P. et al. An automated multi-class skin lesion diagnosis by embedding local and global features of Dermoscopy images. Multimed Tools Appl 82, 34885–34912 (2023). https://doi.org/10.1007/s11042-023-14892-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14892-2