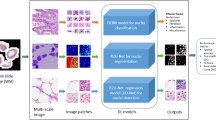
Abstract
Image processing techniques and algorithms are extensively used for biomedical applications. Convolution Neural Network (CNN) is gaining popularity in fields such as the analysis of complex documents and images, which qualifies the approach to be used in biomedical applications. The key drawback of the CNN application is that it can’t call the previous layer output following the layer’s input. To address this issue, the present research has proposed the novel Modified U-Net architecture with ELU Activation Framework (MU-EAF) to detect and classify cancerous cells in the blood smear images. The system is trained with 880 samples, of which 220 samples were utilized in the validation model, and 31 images were utilized to verify the proposed model. The identified mask output of the segmentation model in the predicted mask fits the classification model to identify the cancer cell occurrence in the collected images. In addition, the segmentation evaluation is done by matching each pixel of the ground truth mask (labels) to the predicted labels from the model. The performance metrics for evaluating the segmentation of images are pixel accuracy, dice coefficient (F1-score), and Jaccard coefficient. Moreover, the model is compared with VGG-16 and simple modified CNN models, which have four blocks, each consisting of a convolutional layer, batch normalization, and activation layer with RELU activation function that are implemented and for assessing the same images used for the proposed model. The proposed model shows higher accuracy in comparison.















Similar content being viewed by others
References
Anilkumar K, Manoj V, Sagi T (2021b) Efficacy of cielab and cmyk color spaces in leukemia image analysis: a comparison by statistical techniques. Biomed Eng Appl Basis Commun 33(06):2150042. https://doi.org/10.4015/S1016237221500423
Asuntha A, Srinivasan A (2020) Deep learning for lung cancer detection and classification. Multimed Tools Appl 79(11):7731–7762. https://doi.org/10.1007/s11042-019-08394-3
Bharath B, Kanmani M (2017) Swarm intelligence based image fusion for thermal and visible images. 2017 International Conference on Computation of Power, Energy Information and Commuincation (ICCPEIC), pp 043–048. https://doi.org/10.1109/ICCPEIC.2017.8290336
Carvalho V, Gonçalves IM, Souza A, Souza MS, Bento D, Ribeiro JE, Lima R, Pinho D (2021) Manual and automatic image analysis segmentation methods for blood flow studies in microchannels. Micromachines 12(3):317. https://doi.org/10.3390/mi12030317
Das D, Mahanta LB (2021) A comparative assessment of different approaches of segmentation and classification methods on childhood medulloblastoma images. J Med Biol Eng 41:379–392. https://doi.org/10.1007/s40846-021-00612-4
Elakkiya R, Teja KSS, Deborah LJ, Bisogni C, Medaglia C (2021) Imaging based cervical cancer diagnostics using small object detection - generative adversarial networks. Multimed Tools Appl 81:191–207. https://doi.org/10.1007/s11042-021-10627-3
Elakkiya R, Teja KSS, Deborah LJ, Bisogni C, Medaglia C (2022) Imaging based cervical cancer diagnostics using small object detection - generative adversarial networks. Multimed Tools Appl 81:191–207. https://doi.org/10.1007/s11042-021-10627-3
Ghoneim A, Muhammad G, Hossain MS (2020) Cervical cancer classification using CNNand extreme learning machines. Future Gener Comput Syst 102:643–649. https://doi.org/10.1016/j.future.2019.09.015
Jyothi Priyankaa B, Bhadri Raju MSVS (2021) Machine learning approach for prediction of cervical cancer. Turkish J Comp Math Edu 12(8):3050–3058. https://doi.org/10.17762/turcomat.v12i8.4143
Kalinathan L, Kathavarayan RS, Kanmani M, Dinakaran N (2020) Nuclei detection in hepatocellular carcinoma and dysplastic liver nodules in histopathology images using bootstrap regression. Histol Histopathol 35(10):1115–1123
Kanmani M, Narasimhan V (2019) An optimal weighted averaging fusion strategy for remotely sensed images. Multidim Syst Sign Process 30:1911–1935. https://doi.org/10.1007/s11045-019-00636-9
Kanmani M, Narasimhan V (2020) Optimal fusion aided face recognition from visible and thermal face images. Multimed Tools Appl 79:17859–17883. https://doi.org/10.1007/s11042-020-08628-9
Krithiga R, Geetha P (2020) Deep learning based breast cancer detection and classification using fuzzy merging techniques. Mach Vis Appl 31:63. https://doi.org/10.1007/s00138-020-01122-0
Kumar D, Jain N, Khurana A, Mittal S, Satapathy SC, Senkerik R, Hemanth JD (2020) Automatic detection of white blood cancer from bone marrow microscopic images using convolutional neural networks. IEEE Access 8:142521–142531. https://doi.org/10.1109/ACCESS.2020.3012292
Kurmi Y, Chaurasia V, Kapoor N (2021) Histopathology image segmentation and classification for cancer revelation. Signal Image Video Process 15:1341–1349. https://doi.org/10.1007/s11760-021-01865-x
Li Y, Zhu R, Mi L, Cao Y, Yao D (2016) Segmentation of white blood cell from acute lymphoblastic leukemia images using dual-threshold method. Comput Math Methods Med 2016:1–12. https://doi.org/10.1155/2016/9514707
Madheswari K, Venkateswaran N (2017) Swarm intelligence based optimisation in thermal image fusion using dual tree discrete wavelet transform. Quant InfraRed Thermogr J 14(1):24–43. https://doi.org/10.1080/17686733.2016.1229328
Madheswari K, Venkateswaran N, Sowmiya V (2016) Visible and thermal image fusion using curvelet transform and brain storm optimization. 2016 IEEE region 10 conference (TENCON), pp 2826–2829. https://doi.org/10.1109/TENCON.2016.7848558
Marzahl C, Aubreville M, Voigt J, Maier A (2019) Classification of leukemic B-lymphoblast cells from blood smear microscopic images with an attention-based deep learning method and advanced augmentation techniques. ISBI 2019 C-NMC challenge: classification in cancer cell imaging. Lecture Notes in Bioengineering, Springer, Singapore https://doi.org/10.1007/978-981-15-0798-4_2
Mohammed ZF, Abdulla AA (2021) An efficient CAD system for ALL cell identification from microscopic blood images. Multimed Tools Appl 80:6355–6368. https://doi.org/10.1007/s11042-020-10066-6
Pan X, Yang D, Li L, Liu Z, Yang H, Cao Z, He Y, Ma Z, Chen Y (2018) Cell detection in pathology and microscopy images with multi-scale fully convolutional neural networks. World Wide Web 21(6):1721–1743. https://doi.org/10.1007/s11280-017-0520-7
Ratley A, Minj J, Patre P (2020) Leukemia disease detection and Classification using machine learning approaches: a review. In 2020 First international conference on power, control and computing technologies (ICPC2T), IEEE, pp 161–165. https://doi.org/10.1109/ICPC2T48082.2020.9071471
Ravichandran A, Raja A, Kanmani M (2017) Entropy optimized image fusion: using particle swarm technology and discrete wavelet transform. 2017 International Conference on Computation of Power, Energy Information and Commuincation (ICCPEIC), pp 068–074. https://doi.org/10.1109/ICCPEIC.2017.8290341
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, Springer, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Sahlol AT, Abdeldaim AM, Hassanien AE (2019) Automatic acute lymphoblastic leukemia classification model using social spider optimization algorithm. Soft Comput 23(15):6345–6360. https://doi.org/10.1007/s00500-018-3288-5
Shah S, Nawaz W, Jalil B, Khan HA (2019) Classification of normal and leukemic blast cells in b-all cancer using a combination of convolutional and recurrent neural networks. In ISBI 2019 C-NMC challenge: classification in cancer cell imaging, Springer, pp 23–31. https://doi.org/10.1007/978-981-15-0798-4_3
Shemona JS, Chellappan AK (2020) Segmentation techniques for early cancer detection in red blood cells with deep learning-based classifier—a comparative approach<? show [AQ="" ID=" Q1]"? IET Image Process 14(9):1726–1732. https://doi.org/10.1049/iet-ipr.2019.1067
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):1–48. https://doi.org/10.1186/s40537-019-0197-0
Xiang Y, Sun W, Pan C, Yan M, Yin Z, Liang Y (2020) A novel automation-assisted cervical cancer reading method based on convolutional neural network. Biocybern Biomed Eng 40(2):611–623. https://doi.org/10.1016/j.bbe.2020.01.016
Zeng Z, Xie W, Zhang Y, Lu Y (2019) Ric-unet: an improved neural network based on unet for nuclei segmentation in histology images. IEEE Access 7:21420–21428. https://doi.org/10.1109/ACCESS.2019.2896920
Zheng J, Lin D, Gao Z, Wang S, He M, Fan J (2020) Deep learning assisted efficient AdaBoost algorithm for breast cancer detection and early diagnosis. IEEE Access 8:96946–96954. https://doi.org/10.1109/ACCESS.2020.2993536
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Not Applicable.
Informed consent
Not Applicable.
Disclosure of potential conflict of interest
The authors declare that they have no potential conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Devi, T.G., Patil, N., Rai, S. et al. Real-time microscopy image-based segmentation and classification models for cancer cell detection. Multimed Tools Appl 82, 35969–35994 (2023). https://doi.org/10.1007/s11042-023-14898-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-14898-w