Abstract
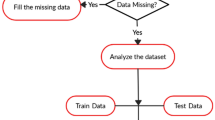
Medical prediction models have gained increasing prevalence in recent years due to their potential to enhance patient outcomes, improve healthcare efficiency, and advance public health. Feature selection and missing data imputation play a key role in medical prediction models. This study aims to analyze the effect of the missing data imputation and filter-based feature selection methods combination on medical prediction models to make a general judgment. We use the four well-known missing data imputation methods (K-Nearest Neighbor, Soft-Impute, Multivariate Imputation by Chained Equations (MICE), and Mean), six commonly used filter-based feature selection methods (Fisher Score, Gini Index, Relieff, Chi-square, Random Forest, and Mutual Information) and three different classifiers (K-Nearest Neighbor: KNN, Logistic Regression: LR, and Support Vector Machine: SVM). We perform all combinations of these models on 6 medical datasets in our experiments. According to Friedman statistical test, which combination of missing data imputation and filter-based feature selection methods used did not affect the performance of medical prediction models where LR and SVM classifiers were used. However, Mean & Chi-square, Mean & GiniIndex combinations statistically perform better than SoftImpute & Fisher score combination for the KNN classifier according to Nemenyi post-hoc statistical test. In addition to these findings, our experiments show that Chi-square has the lowest feature selection run time, while the Relieff method has the longest run time. Besides, we show that all classifiers’ prediction success with feature selection is better than or equal to without feature selection.






Similar content being viewed by others
References
Abdulla M, Khasawneh MT (2022) Integration of aggressive bound tightening and mixed integer programming for cost-sensitive feature selection in medical diagnosis. Expert Syst Appl 187(115):902. https://doi.org/10.1016/j.eswa.2021.115902
Alhassan AM, Wan Zainon WMN (2021) Review of feature selection, dimensionality reduction and classification for chronic disease diagnosis. IEEE Access 9:87310–87317. https://doi.org/10.1109/ACCESS.2021.3088613
Azur MJ, Stuart EA, Frangakis C et al (2011) Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res 20(1):40–49. https://doi.org/10.1002/mpr.329
Battiti R (1994) Using mutual information for selecting features in supervised neural net learning. IEEE Trans Neural Networks 5(4):537–550. https://doi.org/10.1109/72.298224
Bommert A, Sun X, Bischl B et al (2020) Benchmark for filter methods for feature selection in high-dimensional classification data. Comput Stat Data Anal 143(106):839. https://doi.org/10.1016/j.csda.2019.106839
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28. https://doi.org/10.1016/j.compeleceng.2013.11.024
Colombelli F, Kowalski TW, Recamonde-Mendoza M (2022) A hybrid ensemble feature selection design for candidate biomarkers discovery from transcriptome profiles. Knowl-Based Syst 254:109655. https://doi.org/10.1016/j.knosys.2022.109655
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297. https://doi.org/10.1007/bf00994018
Demšar J (2006) Statistical comparisons of classifiers over multiple data sets. Tech Rep. https://doi.org/10.5555/1248547.1248548
Doquire G, Verleysen M (2012) Feature selection with missing data using mutual information estimators. Neurocomputing 90:3–11. https://doi.org/10.1016/j.neucom.2012.02.031
Dua D, Graff C (2017) UCI machine learning repository. http://archive.ics.uci.edu/ml
Elter M, Schulz-Wendtland R, Wittenberg T (2007) The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process. Med Phys 34(11):4164–4172. https://doi.org/10.1118/1.2786864
Fernandes K, Cardoso JS, Fernandes J (2017) Transfer learning with partial observability applied to cervical cancer screening. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 10255 LNCS. Springer Verlag, pp 243–250. https://doi.org/10.1007/978-3-319-58838-4_27
Graham JW (2009). Missing data analysis: making it work in the real world. https://doi.org/10.1146/annurev.psych.58.110405.085530
Hapfelmeier A, Ulm K (2014) Variable selection by Random Forests using data with missing values. Comput Stat Data Anal 80:129–139. https://doi.org/10.1016/j.csda.2014.06.017
He X, Cai D, Niyogi P (2005) Laplacian score for feature selection. In: Proceedings of Advances in Neural Information Processing Systems, pp 507–514
Hu Z, Melton GB, Arsoniadis EG et al (2017) Strategies for handling missing clinical data for automated surgical site infection detection from the electronic health record. J Biomed Inform 68:112–120. https://doi.org/10.1016/j.jbi.2017.03.009
Kira K, Rendell LA (1992) A practical approach to feature selection. In: Machine Learning Proceedings 1992. Elsevier, p 249–256. https://doi.org/10.1016/b978-1-55860-247-2.50037-1
Kononenko I (1994) Estimating attributes: analysis and extensions of RELIEF. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 784 LNCS. Springer Verlag, pp 171–182. https://doi.org/10.1007/3-540-57868-4_57
Lee CH, Yoon HJ (2017) Medical big data: promise and challenges. Kidney Research and Clinical Practice 36(1):3–11. https://doi.org/10.23876/j.krcp.2017.36.1.3
Lee IH, Lushington GH, Visvanathan M (2011) A filter-based feature selection approach for identifying potential biomarkers for lung cancer. Journal of clinical Bioinformatics 1(1):1–8
Little RJA, Rubin DB (2002) Statistical analysis with missing data. Wiley, Hoboken. https://doi.org/10.1002/9781119013563
Liu CH, Tsai CF, Sue KL et al (2020) The feature selection effect on missing value imputation of medical datasets. Appl Sci 10(7):2344. https://doi.org/10.3390/app10072344
Maniruzzaman M, Rahman MJ, Al-MehediHasan M et al (2018) Accurate diabetes risk stratification using machine learning: role of missing value and outliers. J Med Syst 42(5):1–17. https://doi.org/10.1007/s10916-018-0940-7
Mazumder R, Hastie T, Tibshirani R (2010) Spectral regularization algorithms for learning large incomplete matrices. J Mach Learn Res 11(80):2287–2322
Naheed N, Shaheen M, Khan SA et al (2020) Importance of features selection, attributes selection, challenges and future directions for medical imaging data: a review. Computer Modeling in Engineering & Sciences 125(1):314–344
Nematzadeh H, García-Nieto J, Navas-Delgado I et al (2022) Automatic frequency-based feature selection using discrete weighted evolution strategy. Appl Soft Comput 130(109):699. https://doi.org/10.1016/j.asoc.2022.109699
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12(85):2825–2830
Remeseiro B, Bolon-Canedo V (2019) A review of feature selection methods in medical applications. Comput Biol Med 112(103):375. https://doi.org/10.1016/j.compbiomed.2019.103375
Ren K, Fang W, Qu J et al (2020) Comparison of eight filter-based feature selection methods for monthly streamflow forecasting - three case studies on CAMELS data sets. J Hydrol 586(124):897. https://doi.org/10.1016/j.jhydrol.2020.124897
Rubinsteyn A, Feldman S (2016) fancyimpute: an imputation library for python. https://github.com/iskandr/fancyimpute
Sánchez-Maroño N, Alonso-Betanzos A, Tombilla-Sanromán M (2007) Filter methods for feature selection - A comparative study. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 4881 LNCS. Springer Verlag, pp 178–187. https://doi.org/10.1007/978-3-540-77226-2_19
Shiva Darshan SL, Jaidhar CD (2018) Performance evaluation of filter-based feature selection techniques in classifying portable executable files. In: Procedia Computer Science, vol 125. Elsevier B.V., pp 346–356, https://doi.org/10.1016/j.procs.2017.12.046
Solorio-Fernández S, Ariel Carrasco-Ochoa J, Martínez-Trinidad JF (2020) A systematic evaluation of filter unsupervised feature selection methods. Expert Syst Appl 162(113):745. https://doi.org/10.1016/j.eswa.2020.113745
Stiglic G, Kocbek P, Fijacko N et al (2019) Challenges associated with missing data in electronic health records: a case study of a risk prediction model for diabetes using data from Slovenian primary care. Health Informatics Journal 25(3):951–959. https://doi.org/10.1177/1460458217733288
Tang C, Liu X, Zhu X et al (2020) Feature selective projection with low-rank embedding and dual laplacian regularization. IEEE Trans Knowl Data Eng 32(9):1747–1760. https://doi.org/10.1109/TKDE.2019.2911946
Urbanowicz RJ, Meeker M, La Cava W et al (2018). Relief-based feature selection: introduction and review. https://doi.org/10.1016/j.jbi.2018.07.014
van Buuren S (2012) Flexible imputation of missing data. Chapman and Hall/CRC. https://doi.org/10.1201/b11826
van Buuren S, Groothuis-Oudshoorn K (2011) mice: Multivariate imputation by chained equations in R. Journal of Statistical Software 45(3):1–67. https://doi.org/10.18637/jss.v045.i03
Witten IH, Frank E, Hall MA, et al (2016) Data mining: practical machine learning tools and techniques. Elsevier Inc., https://doi.org/10.1016/c2009-0-19715-5
Acknowledgements
This study is supported by Eskisehir Technical University Scientific Research Projects Committee (ESTUBAP-20DRP025).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ergul Aydin, Z., Kamisli Ozturk, Z. Filter-based feature selection methods in the presence of missing data for medical prediction models. Multimed Tools Appl 83, 24187–24216 (2024). https://doi.org/10.1007/s11042-023-15917-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15917-6