Abstract
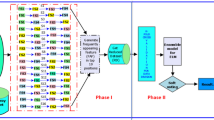
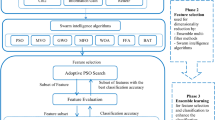
Analysis of microarray gene expression data for the detection/classification of cancer is one of the common approaches adopted worldwide. However, many genes (features) with correlated and irrelevant information in these data sets become the bottleneck for a classification model and significantly deteriorate its performance. A large number of features with fewer samples further make the classification task more cumbersome. Several feature selection methods (both filter and wrapper) have been proposed individually to address this issue, but choosing the best one among them is an open challenge. Our objective in the present study is to simplify the search for the best feature selection method without relying completely on individual methods and propose a two-step hybrid approach. In the first step, we use an ensemble of filter-based heterogeneous feature selection methods. These selected features then undergo the second step of wrapper-based selection. We propose to use the bio-inspired method called Moth-flame optimization (MFO) with an extreme learning machine (ELM) as its fitness function in this step. The motivation for using ELM is to leverage its learning strategy with one-pass processing of samples. Using this hybrid feature selection method, we proposed a classification model for Cancer Micraoarray data, where ELM is also considered as a classifier. The work demonstrates the superiority of the proposed model over other state-of-the-art methods in classifying cancer data from four different microarray gene expression datasets. Several measurement indexes are used for the performance evaluation of models.












Similar content being viewed by others
Data availability
The datasets analysed during the current study are available in the [Mendeley] repository for Lung cancer, Brain cancer and Prostate cancer datasets [https://data.mendeley.com/datasets/ynp2tst2hh/4]
Colon cancer [http://csse.szu.edu.cn/staff/zhuzx/Datasets.html]
References
Ab Hamid TMT, Sallehuddin R, Yunos ZM, Ali A (2021) Ensemble based filter feature selection with harmonize particle swarm optimization and support vector machine for optimal cancer classification. Mach Learn Appl 5:100054
Abdulla M, Khasawneh MT (2020) G-Forest: an ensemble method for cost-sensitive feature selection in gene expression microarrays. Artif Intell Med 108:101941
Alomari OA, Makhadmeh SN, Al-Betar MA, Alyasseri ZAA, Doush IA, Abasi AK, … Zitar RA (2021) Gene selection for microarray data classification based on gray wolf optimizer enhanced with TRIZ-inspired operators. Knowl-Based Syst 223:107034
Alzaqebah M, Alrefai N, Ahmed EA, Jawarneh S, Alsmadi MK (2020) Neighborhood search methods with moth optimization algorithm as a wrapper method for feature selection problems. Int J Electr Comput Eng 10(4):3672
Arowolo MO, Abdulsalam SO, Saheed YK, Salawu MD (2016) A feature selection based on One-Way-Anova for microarray data classification. AJPAS J 3:1–6
Bishop CM (2016) Pattern recognition and machine learning. Springer
Bolón-Canedo V, Alonso-Betanzos A (2019) Ensembles for feature selection: a review and future trends. Inf Fusion 52:1–12
Cao J, Lin Z (2015) Extreme learning machines on high dimensional and large data applications: a survey. Math Probl Eng 2015:1–13
Chuang LY, Yang CH, Wu KC, Yang CH (2011) A hybrid feature selec-tion method for DNA microarray data. Comput Biol Med 41(4):228–237
Dabba A, Tari A, Meftali S (2021) Hybridization of moth flame optimization algorithm and quantum computing for gene selection in microarray data. J Ambient Intell Humaniz Comput 12(2):2731–2750
Dabba A, Tari A, Meftali S, Mokhtari R (2021) Gene selection and classification of microarray data method based on mutual information and moth flame algorithm. Expert Syst Appl 166:114012
Ding Y, Wilkins D (2006) Improving the performance of SVM-RFE to select genes in microarray data. BMC Bioinformatics 7(2):1–8, BioMed Central
Halim Z (2021) An ensemble filter-based heuristic approach for cancerous gene expression classification. Knowl-Based Syst 234:107560
Haznedar B, Arslan M T, Kalinli A (2017) Microarray gene expression cancer data. Mendeley Data, 2
Jackson LA, Dyer DW (2012) Protocol for gene expression profiling using DNA microarrays in Neisseria gonorrhoeae. In: Diagnosis of sexually transmitted diseases. Humana Press, Totowa, NJ, pp 3343–3357
Khurma RA, Aljarah IB, Sharieh AH (2005) Improved moth flame optimization based on Harris hawks for genesselection. J Theoret Appl Inf Technol 98:3794–3807
Kohavi R, John GH (1997) Wrapper feature subset selection. Artif Intell 97(1-2):273–324
Lazar C, Taminau J, Meganck S, Steenhoff D, Coletta A, Molter C, … Nowe A (2012) A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Trans Comput Biol Bioinform 9(4):1106–1119
Li C, Xu J (2019) Feature selection with the fisher score followed by the maximal clique centrality algorithm can accurately identify the hub genes of hepatocellular carcinoma. Sci Rep 9(1):1–11
Li GZ, Zeng XQ, Yang JY, Yang MQ (2007) Partial least squares-based dimension reduction with gene selection for tumour classification. In: 2007 IEEE 7th international symposium on bioinformatics and bioengineering, pp 1439–1444
Li Y, Zhu X, Liu J (2020) An improved moth-flame optimization algorithm for engineering problems. Symmetry 12(8):1234
Liang S, Ma A, Yang S, Wang Y, Ma Q (2018) A review of matched-pairs feature selection methods for gene expression data analysis. Comput Struct Biotechnol 16:88–97
Lin GQ, Li LL, Tseng ML, Liu HM, Yuan DD, Tan RR (2020) An improved moth-flame optimization algorithm for support vector machine prediction of photovoltaic power generation. J Clean Prod 253:119966
Mirjalili S (2015) Moth-flame optimization algorithm: a novel nature-inspired heuristic paradigm. Knowl-Based Syst 89:228–249
Mollaee M, Moattar MH (2016) A novel feature extraction approach based on ensemble feature selection and modified discriminant independent component analysis for microarray data classification. Biocybern Biomed Eng 36(3):521–529
Muduli D, Dash R, Majhi B (2020) Automated breast cancer detection in digital mammograms: a moth flame optimization based ELM approach. Biomedical Signal Processing and Control 59:101912
Nadimi-Shahraki MH, Banaie-Dezfouli M, Zamani H, Taghian S, Mirjalili S (2021) B-MFO: a binary moth-flame optimization for feature selection from medical datasets. Computers 10(11):136
Pashaei E, Ozen M, Aydin N (2016) Gene selection and classification approach for microarray data based on random forest ranking and BBHA. In: 2016 IEEE-EMBS international conference on biomedical and health informatics (BHI). IEEE, pp 308–311
Prakash J, Kankar PK (2020) Health prediction of hydraulic cooling circuit using deep neural network with ensemble feature ranking technique. Measurement 151:107225
Radovic M, Ghalwash M, Filipovic N, Obradovic Z (2017) Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinforma 18(1):1–14
Saeys Y, Inza I, Larranaga P (2007) A review of feature selection techniques in bioinformatics. bioinformatics 23(19):2507–2517
Santos V, Datia N, Pato MPM (2014) Ensemble feature ranking applied to medical data. Proc Technol 17(2014):223–230
Singh N, Singh P (2021) A hybrid ensemble-filter wrapper feature se-lection approach for medical data classification. Chemom Intell Lab Syst 217:104396
Wang M, Chen H, Yang B, Zhao X, Hu L, Cai Z, Tong C (2017) Toward an optimal kernel extreme learning machine using a chaotic moth-flame op-timization strategy with applications in medical diagnoses. Neurocomputing 267:69–84
Wang A, Liu H, Yang J, Chen G (2022) Ensemble feature selection for stable biomarker identification and cancer classification from microarray expression data. Comput Biol Med:105208
Wolpert DH, Macready WG (1997) No free lunch theorems for optimization. IEEE Trans Evol Comput 1:67–82
Zhang G, Hou J, Wang J, Yan C, Luo J (2020) Feature selection for microarray data classification using hybrid information gain and a modified binary krill herd algorithm. Interdiscip Sci Comput Life Sci 12(3):288–301
Zhu Z, Ong YS, Dash M (2007) Markov blanket-embedded genetic algorithm for gene selection. Pattern Recogn 40(11):3236–3248
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sucharita, S., Sahu, B., Swarnkar, T. et al. Classification of cancer microarray data using a two-step feature selection framework with moth-flame optimization and extreme learning machine. Multimed Tools Appl 83, 21319–21346 (2024). https://doi.org/10.1007/s11042-023-16353-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16353-2