Abstract
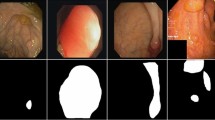
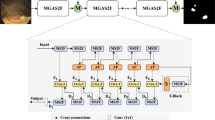
Accurate polyp segmentation is crucial for diagnosing colorectal cancer, but it remains challenging due to shape, texture, and scale variations, as well as difficulties in determining boundaries. Existing methods incorporating attention mechanisms have improved accuracy but lack effective fusion of different level features. Besides, extracting boundary information from surrounding mucosa and early polyps poses challenges. To tackle these issues, a multi-cascade attention-based network (MCA-Net) is proposed. Three components are introduced, including an axial receptive module (ARM), a multi-cascade feature aggregation module (MFA), and an edge fusion module (EFM). The ARM enhances multi-scale analysis by incorporating receptive fields and axial attention, providing the algorithm with a better knowledge of the features. Along with the integration of multi-cascade supervision, MFA selectively refines the information, effectively fusing relevant cues from different levels, suppressing background noise, and highlighting essential polyp features. The EFM focuses on capturing object boundary details, resulting in well-defined and accurate segmentation. Experiment results on five polyp datasets show that our MCA-Net outperforms state-of-the-art (SOTA) methods. Specifically, our MCA-Net achieves an 8.2% improvement in mean Dice compared to the state-of-the-art on the ETIS dataset.







Similar content being viewed by others
Data Availability
The datasets generated during and analysed during the current study are available from the corresponding author on reasonable request.
References
Bhatti UA, Huang M, Wu D, Zhang Y, Mehmood A, Han H (2019) Recommendation system using feature extraction and pattern recognition in clinical care systems. Enterp Inf Syst 13(3):329–351
Nawaz SA, Li J, Bhatti UA, Bazai SU, Zafar A, Bhatti MA, Mehmood A, Ain Qu, Shoukat MU (2021) A hybrid approach to forecast the covid-19 epidemic trend. Plos One 16(10):0256971
Zeeshan Z, Bhatti UA, Memon WH, Ali S, Nawaz SA, Nizamani MM, Mehmood A, Bhatti MA, Shoukat MU et al (2021) Feature-based multi-criteria recommendation system using a weighted approach with ranking correlation. Intell Data Analysis 25(4):1013–1029
Bhatti UA, Yuan L, Yu Z, Nawaz SA, Mehmood A, Bhatti MA, Nizamani MM, Xiao S et al (2021) Predictive data modeling using sp-knn for risk factor evaluation in urban demographical healthcare data. J Med Imaging Health Informatics 11(1):7–14
Ahmad RM, Yao X, Nawaz SA, Bhatti UA, Mehmood A, Bhatti MA, Shaukat MU (2020) Robust image watermarking method in wavelet domain based on sift features. In: Proceedings of the 2020 3rd international conference on artificial intelligence and pattern recognition, pp 180–185
Bhatti UA, Yan Y, Zhou M, Ali S, Hussain A, Qingsong H, Yu Z, Yuan L (2021) Time series analysis and forecasting of air pollution particulate matter (pm 2.5): an sarima and factor analysis approach. IEEE Access 9:41019–41031
Bhatti UA, Zeeshan Z, Nizamani MM, Bazai S, Yu Z, Yuan L (2022) Assessing the change of ambient air quality patterns in jiangsu province of china pre-to post-covid-19. Chemosphere 288:132569
Vinson KE, George DC, Fender AW, Bertrand FE, Sigounas G (2016) The n otch pathway in colorectal cancer. Int J Cancer 138(8):1835–1842. https://doi.org/10.1002/ijc.29800
Magaji BA, Moy FM, Roslani AC, Law CW (2017) Survival rates and predictors of survival among colorectal cancer patients in a malaysian tertiary hospital. BMC Cancer 17(1):1–8. https://doi.org/10.1186/s12885-017-3336-z
Cheng M, Kong Z, Song G, Tian Y, Liang Y, Chen J (2021) Learnable oriented-derivative network for polyp segmentation. In: Medical image computing and computer assisted intervention— MICCAI 2021, pp 720–730 Springer, Cham. https://doi.org/10.1007/978-3-030-87193-2_68
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention — MICCAI 2015, pp 234–241 Springer, Cham. https://doi.org/10.1007/978-3-319-24574-4_28
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. Preprint at https://arxiv.org/abs/1807.10165
Yang X, Li X, Ye Y, Lau RYK, Zhang X, Huang X (2019) Road detection and centerline extraction via deep recurrent convolutional neural network u-net. IEEE Trans Geosci Remote Sens 57(9):7209–7220. https://doi.org/10.1109/TGRS.2019.2912301
Fang Y, Chen C, Yuan Y, K-y Tong, (2019) Selective feature aggregation network with area-boundary constraints for polyp segmentation.In: Medical image computing and computer assisted intervention - MICCAI, (2019) pp 302–310 Springer. Cham. https://doi.org/10.1007/978-3-030-32239-7_34
Fan D-P, Ji G-P, Zhou T, Chen G, Fu H, Shen J, Shao L (2020) Pranet: Parallel reverse attention network for polyp segmentation. In: Medical image computing and computer assisted intervention – MICCAI 2020, pp 263–273 Springer, Cham. https://doi.org/10.1007/978-3-030-59725-2_26
Huang C, Wu H, Lin Y (2021) Hardnet-mseg: a simple encoder-decoder polyp segmentation neural network that achieves over 0.9 mean dice and 86 FPS. Preprint at arXiv:2101.07172
Silva J, Histace A, Romain O, Dray X, Granado B (2014) Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int J CARS 9(2):283–293. https://doi.org/10.1007/s11548-013-0926-3
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F (2015) Wm-dova maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput Med Imaging Graph 43:99–111. https://doi.org/10.1016/j.compmedimag.2015.02.007
Tajbakhsh N, Gurudu SR, Liang J (2016) Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imaging (TMI) 35(2):630–644. https://doi.org/10.1109/TMI.2015.2487997
Vázquez D, Bernal J, Sánchez FJ, Fernández-Esparrach G, López AM, Romero A, Drozdzal M, Courville A (2017) A benchmark for endoluminal scene segmentation of colonoscopy images. Journal of healthcare engineering 2017. https://doi.org/10.1155/2017/4037190
Jha D, Smedsrud PH, Riegler MA, Halvorsen P, de Lange T, Johansen D, Johansen HD (2020) Kvasir-seg: A segmented polyp dataset. In: MultiMedia Modeling, pp 451–462. Springer, Cham. https://doi.org/10.1007/978-3-030-37734-2_37
Jha D, Smedsrud PH, Riegler MA, Johansen D, Lange TD, Halvorsen P, Johansen HD (2019) Resunet++: An advanced architecture for medical image segmentation. In: 2019 IEEE international symposium on multimedia (ISM), pp 225–2255. https://doi.org/10.1109/ISM46123.2019.00049
Ibtehaz N, Rahman MS (2020) Multiresunet : Rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw 121:74–87. https://doi.org/10.1016/j.neunet.2019.08.025
Valanarasu JMJ, Sindagi VA, Hacihaliloglu I, Patel VM (2020) Kiu-net: Towards accurate segmentation of biomedical images using over-complete representations. In: Medical image computing and computer assisted intervention – MICCAI 2020, pp 363–373 Springer, Cham. https://doi.org/10.1007/978-3-030-59719-1_36
Zhang R, Li G, Li Z, Cui S, Qian D, Yu Y (2020) Adaptive context selection for polyp segmentation. In: Medical image computing and computer assisted intervention – MICCAI 2020, pp 253–262 Springer, Cham. https://doi.org/10.1007/978-3-030-59725-2_25
Yin Z, Liang K, Ma Z, Guo J (2022) Duplex contextual relation network for polyp segmentation. In: 2022 IEEE 19th international symposium on biomedical imaging (ISBI), pp 1–5 IEEE. https://doi.org/10.1109/ISBI52829.2022.9761402
Patel K, Bur AM, Wang G (2021) Enhanced u-net: A feature enhancement network for polyp segmentation. In: 2021 18th conference on robots and vision (CRV), pp 181–188 IEEE. https://doi.org/10.1109/CRV52889.2021.00032
Mnih V, Heess N, Graves A et al (2014) Recurrent models of visual attention. Adv Neural Inf Process Syst 27
Xiao T, Xu Y, Yang K, Zhang J, Peng Y, Zhang Z (2015) The application of two-level attention models in deep convolutional neural network for fine-grained image classification. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR), pp 842–850. https://doi.org/10.1109/cvpr.2015.7298685
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition(CVPR), pp 7132–7141. https://doi.org/10.1109/cvpr.2018.00745
Woo S, Park J, Lee J-Y, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19 Springer, Cham. https://doi.org/10.1007/978-3-030-01234-2_1
Zhang H, Goodfellow I, Metaxas D, Odena A (2019) Self-attention generative adversarial networks. In: Proceedings of the 36th international conference on machine learning, vol 97, pp 7354–7363
Sun J, Darbehani F, Zaidi M, Wang B (2020) Saunet: Shape attentive u-net for interpretable medical image segmentation. In: Medical image computing and computer assisted intervention – MICCAI 2020, pp 797–806 Springer, Cham. https://doi.org/10.1007/978-3-030-59719-1_77
Ho J, Kalchbrenner N, Weissenborn D, Salimans T (2019) Axial attention in multidimensional transformers. Preprint at arXiv:1912.12180
Chu X, Tian Z, Wang Y, Zhang B, Ren H, Wei X, Xia H, Shen C (2021) Twins: Revisiting the design of spatial attention in vision transformers. Adv Neural Inf Process Syst 34:9355–9366
Wang H, Zhu Y, Green B, Adam H, Yuille A, Chen L-C (2020) Axial-deeplab: Stand-alone axial-attention for panoptic segmentation. In: European conference on computer vision – ECCV 2020, pp 108–126 Springer, Cham. https://doi.org/10.1007/978-3-030-58548-8_7
Wang P, Chen P, Yuan Y, Liu D, Huang Z, Hou X, Cottrell G (2018) Understanding convolution for semantic segmentation. In: 2018 IEEE winter conference on applications of computer vision (WACV), pp 1451–1460 IEEE. https://doi.org/10.1109/WACV.2018.00163
Zhao X, Zhang L, Lu H (2021) Automatic polyp segmentation via multi-scale subtraction network. In: Medical image computing and computer assisted intervention – MICCAI 2021, pp 120–130 Springer, Cham. https://doi.org/10.1007/978-3-030-87193-2_12
Ioffe S, Szegedy C (2015) Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning, pp 448–456 PMLR
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics, vol 15, pp 315–323 PMLR
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. Preprint at arXiv:1412.6980
Achanta R, Hemami S, Estrada F, Susstrunk S (2009) Frequency-tuned salient region detection. In: 2009 IEEE conference on computer vision and pattern recognition(CVPR), pp 1597–1604. https://doi.org/10.1109/CVPR.2009.5206596
Fan D-P, Cheng M-M, Liu Y, Li T, Borji A (2017) Structure-measure: A new way to evaluate foreground maps. In: Proceedings of the IEEE international conference on computer vision, pp 4548–4557. https://doi.org/10.1109/iccv.2017.487
Fan D-P, Gong C, Cao Y, Ren B, Cheng M-M, Borji A (2018) Enhanced-alignment measure for binary foreground map evaluation. Preprint at arXiv:1805.10421
Perazzi F, Krähenbühl P, Pritch Y, Hornung A (2012) Saliency filters: Contrast based filtering for salient region detection. In: 2012 IEEE conference on computer vision and pattern recognition(CVPR), pp 733–740. https://doi.org/10.1109/CVPR.2012.6247743
Dong B, Wang W, Fan D-P, Li J, Fu H, Shao L (2021) Polyp-pvt: Polyp segmentation with pyramid vision transformers. Preprint at arXiv:2108.06932
Chao P, Kao C-Y, Ruan Y, Huang C-H, Lin Y-L (2019) Hardnet: A low memory traffic network. In: 2019 IEEE/CVF international conference on computer vision (ICCV), pp 3551–3560. https://doi.org/10.1109/ICCV.2019.00365
Acknowledgements
This research is supported by the National Key Research and Development Program of China (2018YFB0804202, 2018YFB0804203), Regional Joint Fund of NSFC (U19A2057), the National Natural Science Foundation of China (61876070), Jilin University “Interdisciplinary Integration and Innovation” Young Scholars Free Exploration Project (JLUXKJC2021QZ01), Jilin Province Science and Technology Development Plan Project (20190303134SF), Anhui University Collaborative Innovation Project Subproject (GXXT-2021-008). The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Shen, X., Lyu, Y. et al. MCA-Net: multi-cascade attention network for polyp segmentation. Multimed Tools Appl 83, 33713–33730 (2024). https://doi.org/10.1007/s11042-023-16805-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16805-9