Abstract
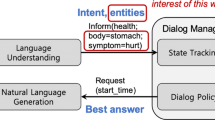
China has entered a stage of high-quality development, and people have higher demand and expectations for the existing medical field. MNER(Medical Named Entity Recognition) is the task of identifying the correct entity boundary and classifying medical entities from a piece of medical text information. The effect of MNER would directly affect the performance of downstream relationship extraction and intelligent question answering, which has important research significance and value. In this paper, aiming at the named entity recognition of discontinuous medical entities in Chinese medical text information, this paper expands the research based on BiLSTM-CRF, introduces the IDCNN layer after the input layer of the model to capture the local context information in the medical text, and then uses the output of IDCNN as the input of BiLSTM-CRF for subsequent training, to construct an IDCNN-BiLSTM-CRF network model for discontinuous medical text. Based on BERT, the weight is assigned to the 12-layer transformer in BERT, and the results are weighted and averaged, and a WfBERT-Att-D-BiLSTM-CRF model based on the weight output of different layers of BERT is proposed. During the experiment, the hidden layer, the number of iterations, and the batch data size are continuously experimented with, and finally, the optimal parameter settings are obtained. In this paper, repeated experiments are carried out on different datasets, the final actual recognition effect is also tested, and the correctness of the proposed model is verified from different perspectives.










Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Zhang X, Liu S, Wang H (2023) Personalized learning path recommendation for e-learning based on knowledge graph and graph convolutional network[J]. Int J Software Eng Knowl Eng 33(01):109–131
Lee LH, Lu Y (2021) Multiple embeddings enhanced multi-graph neural networks for Chinese Healthcare named entity recognition[J]. IEEE J Biomed Health Inform 25(7):2801–2810
Ramachandran R, Arutchelvan K (2021) Grey Wolf optimizer with conditional random fields for healthcare related medical named entity recognition model[J]. SSRN Electron J. https://doi.org/10.2139/ssrn.3769845
Quimbaya AP, Múnera AS, Rivera RA, Rodríguez JC, Velandia OM, Peña AA, Labbé C (2016) Named entity recognition over electronic health records through a combined dictionary-based approach[J]. Procedia Comput Sci 100:55–61. https://doi.org/10.1016/j.procs.2016.09.123
Wang X, Chused A, Elhadad N, Friedman C, Markatou M (2008) Automated knowledge acquisition from clinical narrative reports. In: Proceedings of the 2008 AMIA Annual Symposium, p 783–787
Bhatia S, Sharma M, Bhatia KK (2015) Strategies for mining opinions: A survey. In: Proc. of the 2nd Int'l Conf. on Computing for Sustainable Global Development (INDIACom 2015). Washington: IEEE Computer Society pp 262–266
Rebecka W, Linus H, Agnes T et al (2014) Visual entity linking: A preliminary study[C]. Proceedings of Workshops at the Twenty-Eighth AAAI Conference on Artificial Intelligence(AAAI-14): 46–49
Li L, Zhao J, Hou L et al (2019) An attention-based deep learning model for clinical named entity recognition of Chinese electronic medical records[J]. BMC Med Inform Decis Mak 19(Suppl 5):235. https://doi.org/10.1186/s12911-019-0933-6
Pomares-Quimbaya A, Gonzalez RA, Velandia OMM et al (2018) Concept attribute labeling and context-aware named entity recognition in electronic health records[J]. Int J Reliab Qual E-Healthc (IJRQEH) 7
Wang Q, Zhou Y, Ruan T, Gao D, Xia Y, He P (2019) Incorporating dictionaries into deep neural networks for the Chinese clinical named entity recognition[J]. J Biomed Inform 92
Tay Y, Tuan LA, Hui SC (2018) Multi-cast attention networks for retrieval-based question answering and response prediction[J]. https://doi.org/10.48550/arXiv.1806.00778
Yu F, Koltun V (2016) Multi-scale context aggregation by dilated convolutions[C]//ICLR. https://doi.org/10.48550/arXiv.1511.07122
Yang B, Mitchell T (2019) Leveraging knowledge bases in LSTMs for improving machine reading[J]. https://doi.org/10.48550/arXiv.1902.09091
Yang YS, Zhang M, Chen W et al (2018) Adversarial learning for Chinese NER from crowd annotations. https://doi.org/10.48550/arXiv.1801.05147
Zhang Y, Yang J (2018) Chinese ner using lattice lstm[J]. arXiv preprint arXiv:1805.02023
Xia Y, Wang Q (2017) Clinical named entity recognition: ECUST in the CCKS-2017 shared task 2[C]. CEUR Workshop Proceedings. Chengdu, China: the Technical Committee on Language and Knowledge Computing of The Chinese Information Processing Society of China 1976:43–48
Huang L, Yu C, Chi Y et al (2019) Towards smart healthcare management based on knowledge graph technology[C]. The 2019 8th International Conference
Dong X, Chowdhury S, Qian L et al (2017) Transfer bi-directional lstm rnn for named entity recognition in Chinese electronic medical records[C], 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom). IEEE pp 1–4
Liu M, Zhang Y, Li W et al (2020) Joint model of entity recognition and relation extraction with self-attention mechanism[J]. ACM Transactions on Asian and Low-Resource Language Information Processing (TALLIP). https://doi.org/10.1145/3387634
Xue K, Zhou Y, Ma Z et al (2019) Fine-tuning BERT for joint entity and relation extraction in Chinese medical text. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp 892–897
Wei Q, Zhou Y, Zhao B et al (2020) Named entity recognition from table headers in randomized controlled trial articles[C]. 2020 IEEE International Conference on Healthcare Informatics (ICHI). IEEE
Morales-Burton V, Lopez-Ramirez SA (2022) Rethinking healthcare quality and prestige: is this a manager's number one problem?[J]. LSE Research Online Documents on Economics
Li X, Zhang H, Zhou XH (2020) Chinese clinical named entity recognition with variant neural structures based on BERT methods. J Biomed Inform 107:103422
Wen G, Chen H, Li H et al (2020) Cross domains adversarial learning for Chinese named entity recognition for online medical consultation. J Biomed Inform 112:103608
Li Z, Gan Z, Zhang B et al. Noisy label learning for Chinese medical named entity recognition based on uncertainty strategy. Proceedings of the Evaluation Tasks at the China Conference on Knowledge Graph and Semantic Computing (CCKS 2020)
Xue K, Zhou Y, Ma Z et al (2019) Fine-tuning BERT for joint entity and relation extraction in Chinese medical text[J]. IEEE. https://doi.org/10.1109/BIBM47256.2019.8983370
Yin R, Wang Q, Li P et al (2016) Multi-granularity chinese word embedding[C]. Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing: 981–986
Yin R, Wang Q, Li P et al (2016) Multi-granularity Chinese word embedding[C]//conference on empirical methods in natural language processing. https://doi.org/10.18653/v1/D16-1100
Li Y, Wang X, Hui L et al (2020) Chinese clinical named entity recognition in electronic medical records: Development of a lattice long short-term memory model with contextualized character representations. JMIR Med Inform 8(9):e19848
Landolsi MY, Hlaoua L, Romdhane LB (2022) infonnation extraction from e1ectronic medica1 documents state of the art and future research directions[J]. Knowl Inf Syst 65(2):463–516. https://doi.org/10.1007/s10115-022-01779-1
Landolsi MY, Romdhane LB, Hlaoua L (2022) Medical named entity recognition using surrounding sequences matching. Procedia Comput Sci[J] 207:674–683
Méndez M, Merayo MG, Núñez M (2023) Long-term traffic flow forecasting using a hybrid CNN-BiLSTM model. Eng Appl Artif Intell[J] 121:106041
Sui Y, Bu F, Hu Y et al (2022) Trigger-GNN: A trigger-based graph neural network for nested named entity recognition[J]. https://doi.org/10.1109/IJCNN55064.2022.9892555
Hssayni EH, Joudar NE, Ettaouil M (2022) Localization and reduction of redundancy in CNN using L1-sparsity induction[J]. J Ambient Intell Human Comput: 1–13. https://doi.org/10.1007/s12652-022-04025-2
Hssayni EH, Joudar N-E, Ettaouil M (2022) An adaptive Drop method for deep neural networks regularization: Estimation of DropConnect hyperparameter using generalization gap. Knowl Based Syst [J] 253:109567
Hssayni Eh, Joudar N-E, Ettaouil M (2022) A deep learning framework for time series classification using normal cloud representation and convolutional neural network optimization. Comput Intell 38(6):2056–2074
Cui Y, Che W, Liu T et al (2021) Pre-training with whole wordmasking for chinese bert[J]. IEEE/ACM Trans Audio Speech Lang Process 29:3504–3514
Jin Y, Xiong Y, Shi D et al (2023) Learning from undercodedclinical records for automated International Classification of Diseases (ICD) coding[J]. J Am Med Inform Assoc 30(3):438–446
Li X, Sun Z, Zhu G (2023) CCRFs-NER: Named entity recognition method based on cascaded conditional random fields oriented Chinese EMR[C]. Tenth Inter-national Conference on Applications and Techniques in Cyber Intelli gence (ICATCI 2022) volume 2. Springer International Publishing, Cham, pp 229–237
Cui S, Joe I (2023) A multi-head adjacent attention-based pyramidlayered model for nested named entity recognition[J]. Neural Comput Appl 35(3):2561–2574
Yao L, Jiang MF, Fang X et al (2022) Research on chinese clinical named entity recognition method based on radical feature and BERT-Transformer-CRF[J]. Softw Eng 25(12):30–36. https://doi.org/10.19644/j.cnki.issn2096-1472.2022.012.007
Acknowledgements
This work is supported by the National Natural Science Foundation of China (61872284); Industrial field of general projects of science and Technology Department of Shaanxi Province(2023-YBGY-203, 2023-YBGY-021); Industrialization Project of Shaanxi Provincial Department of Education (21JC017); "Thirteenth Five-Year" National Key R&D Program Project (Project Number: 2019YFD1100901). Natural Science Foundation of Shannxi Province, China(2021JLM-16).
Author information
Authors and Affiliations
Contributions
Qinlu He and Zhen Li contributed significantly to Design Algorithm. Genqing Bian and Zan Wang performed the perfor analysis. Pengze Gao completed the experimental part. Fan Zhang and Qinlu He contributed to manuscript preparation.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The authors declare that they have no conflicts of interest.
Consent for publication
I have read and understood the publishing policy, and submit this manuscript in accordance with this policy.
The results/data/figures in this manuscript have not been published elsewhere, nor are they under consideration (from you or one of your Contributing Authors) by another publisher.
Competing interests
I declare that the authors have no competing interests as defined by Springer, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, Q., Gao, P., Zhang, F. et al. Healthcare entity recognition based on deep learning. Multimed Tools Appl 83, 32739–32763 (2024). https://doi.org/10.1007/s11042-023-16900-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16900-x