Abstract
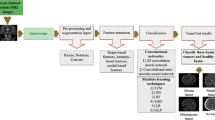
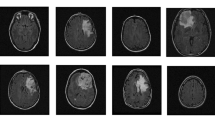
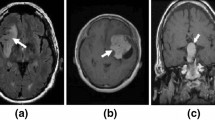
Deep Learning has significantly push forward the research on cancer magnetic resonance imaging (MRI) images. These images are widely used to diagnose the presence of a deformed tissue within the brain in which the cells replicate indefinitely without control, i.e. a brain tumor. Radiologist have to deeply examine a set of MRI images for each patient in order to decide whether the tumor is benign (noncancerous) or malignant (cancerous). The latest have very severe consequences and have a very high mortality rate, but this could be significantly reduced if the cancer is diagnosed at an earlier stage. The classification task is very complicated due to neurological and radiological similarities of different tumors. In order to assist the radiologists, our objective in this paper is to achieve a correct classification of the MRI images. The studied deep classification models have been trained over three types of tumors: meningioma, glioma and pituitary tumor, on sagittal, coronal and axial views in addition to MRI of normal patients. The proposed model consists of combining a set of several classifiers that uses the features extracted by a convolutional neural network (CNN). We will, also, explore the impact of data augmentation and image resolution on the classification performance with the goal of obtaining the best possible accuracy. We used a CNN architecture and several pre-trained models. The model ResNet 18 gave the highest accuracy of 95.7%.







Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Buckner JC, Brown PD, O’Neill BP, Meyer FB, Wetmore CJ, Uhm JH (2007), October Central nervous system tumors. In: Mayo Clinic Proceedings (Vol. 82, No. 10). Elsevier, Amsterdam, pp 1271–1286. https://doi.org/10.4065/82.10.1271
Thangudu S, Cheng FY, Su CH (2020) Advancements in the blood–brain barrier penetrating nanoplatforms for brain related disease diagnostics and therapeutic applications. Polymers 12(12):3055
Komori T (2020) Updating the grading criteria for adult diffuse gliomas: beyond the WHO2016CNS classification. Brain Tumor Pathol 37(1):1–4. https://doi.org/10.1007/s10014-020-00358-y
Koriyama S, Nitta M, Kobayashi T, Muragaki Y, Suzuki A, Maruyama T, Kawamata T (2018) A surgical strategy for lower grade gliomas using intraoperative molecular diagnosis. Brain Tumor Pathol 35:159–167. https://doi.org/10.1007/s10014-018-0324-1
Asano K, Kurose A, Kamataki A, Kato N, Ogawa K, Katayama K, Ohkuma H (2018) Importance and accuracy of intraoperative frozen section diagnosis of the resection margin for effective carmustine wafer implantation. Brain Tumor Pathol 35:131–140. https://doi.org/10.1007/s10014-018-0320-5
Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M (1975) Cronin KA. SEER cancer statistics review, 2015
Ohgaki H, Kleihues P (2005) Epidemiology and etiology of gliomas. Acta Neuropathol 109:93–108. https://doi.org/10.1007/s00401-005-0991-y
Akagi Y, Yoshimoto K, Hata N, Kuga D, Hatae R, Amemiya T, Iihara K (2018) Reclassification of 400 consecutive glioma cases based on the revised 2016WHO classification. Brain Tumor Pathol 35:81–89. https://doi.org/10.1007/s10014-018-0313-4
Kuwahara K, Ohba S, Nakae S, Hattori N, Pareira ES, Yamada S, Hirose Y (2019) Clinical, histopathological, and molecular analyses of IDH-wild-type WHO grade II–III gliomas to establish genetic predictors of poor prognosis. Brain Tumor Pathol 36:135–143. https://doi.org/10.1007/s10014-019-00348-9
Spatharou A, Hieronimus S, Jenkins J (2020) Transforming healthcare with AI: The impact on the workforce and organizations. McKinsey & Company: Sydney, NSW, Australia, vol 10
Jerban S, Chang EY, Du J (2020) Magnetic resonance imaging (MRI) studies of knee joint under mechanical loading. Magn Reson Imaging 65:27–36. https://doi.org/10.1016/j.mri.2019.09.007
Spadaccini M, Iannone A, Maselli R, Badalamenti M, Desai M, Chandrasekar VT, Repici A (2021) Computer-aided detection versus advanced imaging for detection of colorectal neoplasia: a systematic review and network meta-analysis. Lancet Gastroenterol Hepatol 6(10):793–802. https://doi.org/10.1016/S2468-1253(21)00215-6
Saygılı A (2021) A new approach for computer-aided detection of coronavirus (COVID-19) from CT and X-ray images using machine learning methods. Appl Soft Comput 105:107323. https://doi.org/10.1016/j.asoc.2021.107323
Jungblut L, Blüthgen C, Polacin M, Messerli M, Schmidt B, Euler A, Martini K (2022) First performance evaluation of an artificial intelligence-based computer-aided detection system for pulmonary nodule evaluation in dual-source photon-counting detector CT at different low-dose levels. Invest Radiol 57(2):108–114. https://doi.org/10.1097/RLI.0000000000000814
Jarnalo CM, Linsen PVM, Blazís SP, van der Valk PHM, Dieckens DBM (2021) Clinical evaluation of a deep-learning-based computer-aided detection system for the detection of pulmonary nodules in a large teaching hospital. Clin Radiol 76(11):838–845. https://doi.org/10.1016/j.crad.2021.07.012
Alshayeji MH, Ellethy H, Gupta R (2022) Computer-aided detection of breast cancer on the Wisconsin dataset: an artificial neural networks approach. Biomed Signal Process Control 71:103141. https://doi.org/10.1016/j.bspc.2021.103141
Bahadure NB, Ray AK, Thethi HP (2018) Comparative approach of MRI-based brain tumor segmentation and classification using genetic algorithm. J Digit Imaging 31:477–489. https://doi.org/10.1007/s10278-018-0050-6
Singh R, Goel A, Raghuvanshi DK (2021) Computer-aided diagnostic network for brain tumor classification employing modulated Gabor filter banks. Visual Comput 37(8):2157–2171. https://doi.org/10.1007/s00371-020-01977-4
Abdelaziz M, Cherfa Y, Cherfa A, Alim-Ferhat F (2021) Automatic brain tumor segmentation for a computer‐aided diagnosis system. Int J Imaging Syst Technol 31(4):2226–2236. https://doi.org/10.1002/ima.22594
Samanta AK, Khan AA (2018) Computer aided diagnostic system for automatic detection of brain tumor through MRI using clustering based segmentation technique and SVM classifier. In: The International Conference on Advanced Machine Learning Technologies and Applications (AMLTA2018). Springer International Publishing, pp 343–351. https://doi.org/10.1007/978-3-319-74690-6_34
Kleesiek J, Biller A, Urban G, Kothe U, Bendszus M, Hamprecht F (2014) Ilastik for multi-modal brain tumor segmentation. Proceedings MICCAI BraTS (brain tumor segmentation challenge), pp 12–17
Addeh A, Iri M (2021) Brain tumor type classification using deep features of MRI images and optimized RBFNN. ENG Trans 2:1–7
Cruz DPF, Maia RD, da Silva LA, de Castro LN (2016) BeeRBF: a bee-inspired data clustering approach to design RBF neural network classifiers. Neurocomputing 172:427–437
Gu K, Zhang Y, Qiao J (2020) Ensemble meta-learning for few-shot soot density recognition. IEEE Trans Industr Inf 17(3):2261–2270. https://doi.org/10.1109/TII.2020.2991208
Gu K, Liu H, Xia Z, Qiao J, Lin W, Thalmann D (2021) PM2.5 monitoring: use information abundance measurement and wide and deep learning. IEEE Trans Neural Networks Learn Syst 32(10):4278–4290. https://doi.org/10.1109/TNNLS.2021.3105394
Gu K, Xia Z, Qiao J, Lin W (2019) Deep dual-channel neural network for image-based smoke detection. IEEE Trans Multimedia 22(2):311–323. https://doi.org/10.1109/TMM.2019.2929009
Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, … Feng Q (2015) Correction:enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS One 10(12):e0144479. https://doi.org/10.1371/journal.pone.0144479
Hubel DH, Wiesel TN (1968) Receptive fields and functional architecture of monkey striate cortex. J Physiol 195(1):215–243. https://doi.org/10.1113/jphysiol.1968.sp008455
Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6–12, 2014, Proceedings, Part V 13. Springer International Publishing, pp 740–755
Everingham M, Van Gool L, Williams CK, Winn J, Zisserman A (2010) The pascal visual object classes (voc) challenge. Int J Comput Vision 88:303–338. https://doi.org/10.1007/s11263-009-0275-4
Ruder S (2016) An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747. https://doi.org/10.48550/arXiv.1609.04747
Li S, Wang L, Li J, Yao Y (2021) Image classification algorithm based on improved AlexNet. In: Journal of Physics: Conference Series (Vol.1813, No. 1). IOP Publishing, p 012051. https://doi.org/10.1088/1742-6596/1813/1/012051
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, … Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, pp 248–255. https://doi.org/10.1109/CVPR.2009.5206848
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. https://doi.org/10.48550/arXiv.1409.1556
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, … Adam H (2017) Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861. https://doi.org/10.48550/arXiv.1704.04861
Ioffe S, Szegedy C (2015) Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning, PMLR, pp 448–456
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Redmon J, Farhadi A (2018) Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767. https://doi.org/10.48550/arXiv.1804.02767
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All authors designed the study. The author performed the simulations and wrote the manuscript. All authors read, edited, and approved the manuscript.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Benbakreti, S., Benouis, M., Roumane, A. et al. Impact of the data augmentation on the detection of brain tumor from MRI images based on CNN and pretrained models. Multimed Tools Appl 83, 39459–39478 (2024). https://doi.org/10.1007/s11042-023-17092-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-17092-0