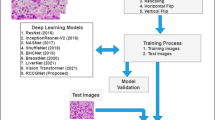
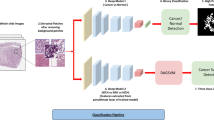
Abstract
Renal Cell Carcinoma (RCC) is the most common malignant tumor (85%) of kidney cancer and has a complex histological pattern and nuclear structure. The manual diagnosis of kidney cancer or any other cancer from histopathology image depends on the knowledge and experience of pathologists, and the pathologist’s experience influences the results. According to studies, the kind of histology in kidney cancer is related to the prognosis and course of treatment. Since the kind of histology, molecular profile, and stage of the disease all affect how the disease is treated, there is an essential need to develop an automated system that can precisely analyze the histopathological images of the disease. This work demonstrates how a deep learning framework can be used to predict and classify associated grades of RCC from provided haematoxylin and eosin (H &E) images. The proposed model focuses on two important tasks- First to capture and extract associated features from the H &E images of five different grades. Second, to classify the new set of unseen H &E images into five separate grades using the obtained features. The proposed architecture has been tested and experimented on two independent datasets containing H &E stained histopathology images. The proposed architecture has been examined using the following performance metrics namely precision, recall, F1 - score, accuracy, Floating-point operations (FLOPs), and the total number of parameters. The obtained results show that the proposed architecture attains better results over seven state-of-the-art deep learning architectures on two different H &E stained histopathology image datasets.





Similar content being viewed by others
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Aatresh AA, Alabhya K, Lal S, Kini J, Saxena PU (2021) Livernet: efficient and robust deep learning model for automatic diagnosis of sub-types of liver hepatocellular carcinoma cancer from h &e stained liver histopathology images. Int J CARS 16(9):1549–1563
Adeshina SA, Adedigba AP, Adeniyi AA, Aibinu AM (2018) Breast cancer histopathology image classification with deep convolutional neural networks. In: 2018 14th international conference on electronics computer and computation (ICECCO), pp 206–212. IEEE
Alom MZ, Yakopcic C, Nasrin M, Taha TM, Asari VK et al (2019) Breast cancer classification from histopathological images with inception recurrent residual convolutional neural network. J Digit Imaging 32(4):605–617
Baranwal N, Doravari P, Kachhoria R (2021) Classification of histopathology images of lung cancer using convolutional neural network (cnn). Disruptive Dev Biomed Appl, p 75
Byeon S, Park J, Cho YA, Cho B-J (2022) Automated histological classification for digital pathology images of colonoscopy specimen via deep learning. Sci Rep 12(1):12804
Chanchal AK, Lal S, Barnwal D, Sinha P, Arvavasu S, Kini J (2023) Evolution of livernet 2. x: Architectures for automated liver cancer grade classification from h &e stained liver histopathological images. Multimed Tools Appl 1–31
Chanchal AK, Lal S, Kumar R, Kwak JT, Kini J (2023) A novel dataset and efficient deep learning framework for automated grading of renal cell carcinoma from kidney histopathology images. Sci Rep 13(5728)
Gupta KD, Sharma DK, Ahmed S, Gupta H, Gupta D, Hsu C-H (2023) A novel lightweight deep learning-based histopathological image classification model for iomt. Neural Process Lett 55(1):205–228
Dogar GM, Shahzad M, Fraz MM (2023) Attention augmented distance regression and classification network for nuclei instance segmentation and type classification in histology images. Biomed Signal Process Control 79:104199
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S et al (2010) An image is worth 16x16 words: transformers for image recognition at scale. arXiv:2010.11929
Eerapu KK, Ashwath B, Lal S, Dell’Acqua F, Dhan AVN (2019) Dense refinement residual network for road extraction from aerial imagery data. IEEE Access 7:151764–151782
Hameed Z, Garcia-Zapirain B, Aguirre JJ, Isaza-Ruget MA (2022) Multiclass classification of breast cancer histopathology images using multilevel features of deep convolutional neural network. Sci Rep 12(1):15600
Han S, Hwang SI, Lee HJ (2019) The classification of renal cancer in 3-phase ct images using a deep learning method. J Digit Imaging 32(4):638–643
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hirra I, Ahmad M, Hussain A, Ashraf MU, Saeed IA, Qadri SF, Alghamdi AM, Alfakeeh AS (2021) Breast cancer classification from histopathological images using patch-based deep learning modeling. IEEE Access 9:24273–24287
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H (2017) Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv:1704.04861
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M (2020) Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci Rep 10(1):1–11
Jiang Y, Chen L, Zhang H, Xiao X (2019) Breast cancer histopathological image classification using convolutional neural networks with small se-resnet module. PloS one 14(3):e0214587
Joseph AA, Abdullahi M, Junaidu SB, Ibrahim HH, Chiroma H (2022) Improved multi-classification of breast cancer histopathological images using handcrafted features and deep neural network (dense layer). Intell Syst Appl 14:200066
Khoshdeli M, Borowsky A, Parvin B (2018) Deep learning models differentiate tumor grades from h &e stained histology sections. In: 2018 40th Annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 620–623. IEEE
Kingma DP, Ba JL (2015) Adam: A method for stochastic optimization. International Conference on Learning Representations ICLR. https://arxiv.org/pdf/1412.6980.pdf
Koidl K (2013) Loss functions in classification tasks. School of Computer Science and Statistic Trinity College, Dublin
Kumar A, Vishwakarma A, Bajaj V (2023) Crccn-net: Automated framework for classification of colorectal tissue using histopathological images. Biomed Signal Process Control 79:104172
Lal S, Desouza R, Maneesh M, Kanfade A, Kumar A, Perayil G, Alabhya K, Chanchal KA, Kini J (2020) A robust method for nuclei segmentation of h &e stained histopathology images. In: 2020 7th International conference on signal processing and integrated networks (SPIN), pp 453–458. IEEE
Mondol RK, Millar EKA, Graham PH, Browne L, Sowmya A, Meijering E (2023) hist2rna: an efficient deep learning architecture to predict gene expression from breast cancer histopathology images. Cancers 15(9):2569
Motlagh MH, Jannesari M, Aboulkheyr H, Khosravi P, Elemento O, Totonchi M, Hajirasouliha I (2018) Breast cancer histopathological image classification: a deep learning approach. BioRxiv, p 242818
Moyes A, Gault R, Zhang K, Ming J, Crookes D, Wang J (2023) Multi-channel auto-encoders for learning domain invariant representations enabling superior classification of histopathology images. Med Image Anal 83:102640
Nahid A-A, Mehrabi MA, Kong Y (2018) Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. BioMed Res Int 2018
Narayanan BN, Krishnaraja V, Ali R (2019) Convolutional neural network for classification of histopathology images for breast cancer detection. In: 2019 IEEE National aerospace and electronics conference (NAECON), pp 291–295. IEEE
Shahidi F, Daud SM, Abas H, Ahmad NA, Maarop N (2020) Breast cancer classification using deep learning approaches and histopathology image: a comparison study. IEEE Access 8:187531–187552
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2016) Breast cancer histopathological image classification using convolutional neural networks. In: 2016 international joint conference on neural networks (IJCNN), pp 2560–2567. IEEE
Srikantamurthy MM, Rallabandi VP, Dudekula DB, Natarajan S, Park J (2023) Classification of benign and malignant subtypes of breast cancer histopathology imaging using hybrid cnn-lstm based transfer learning. BMC Med Imaging 23(1):1–15
Sun C, Xu A, Liu D, Xiong Z, Zhao F, Ding W (2019) Deep learning-based classification of liver cancer histopathology images using only global labels. IEEE J Biomed Health Inf 24(6):1643–1651
Sun K, Chen Y, Bai B, Gao Y, Xiao J, Yu G (2023) Automatic classification of histopathology images across multiple cancers based on heterogeneous transfer learning. Diagnostics 13(7):1277
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Toğaçar M, Özkurt KB, Ergen B, Cömert Z (2020) Breastnet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys A: Stat Mech Appl 545:123592
Wakili MA, Shehu HA, Sharif MH, Sharif MHU, Umar A, Kusetogullari H, Ince IF, Uyaver S et al (2022) Classification of breast cancer histopathological images using densenet and transfer learning. Comput Intell Neurosci 2022
Woo S, Park J, Lee J-Y, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1492–1500
Yan R, Ren F, Wang Z, Wang L, Zhang T, Liu Y, Rao X, Zheng C, Zhang F (2020) Breast cancer histopathological image classification using a hybrid deep neural network. Methods 173:52–60
Zhang X, Zhou X, Lin M, Sun J (2018) Shufflenet: An extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6848–6856
Acknowledgements
The authors would like to thank the handling editor, editor-in-chief, and anonymous reviewers for their valuable suggestions and comments which helped to improve the quality of the research paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
Authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chanchal, A.K., N, S., Lal, S. et al. Classification and grade prediction of kidney cancer histological images using deep learning. Multimed Tools Appl 83, 78247–78267 (2024). https://doi.org/10.1007/s11042-024-18639-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-024-18639-5