Abstract
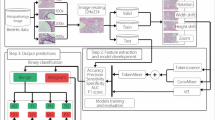
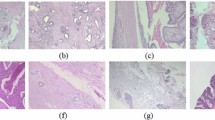
Breast cancer (BC) is a widespread and lethal cancer affecting women world- wide. Early diagnosis plays a pivotal role in ensuring survival, as late detection can result in a fatal outcome. Convolutional neural networks (CNNs) have made significant contributions to the task of medical imaging modalities and have dis- played promise in addressing this challenge. Recently, the success of the vision transformer (ViT) architecture has encouraged the use of the attention mecha- nism in computer-aided diagnosis (CAD) tasks. However, the ViT is known for its data-intensive nature and a substantial number of parameters and needs power- ful computer resources when training, which often leads to the same performance compared to CNNs. These challenges are particularly evident in tasks involving medical image datasets with complex images and limited data. This problem- atic situation led to the suggestion three of low-weight parameter systems based on convolution and attention techniques: vision transformer base model (ViT), compact convolution transformers (CCT), and lightweight mobile vision trans- formers (MVIT). These systems are developed by using the BreakHis dataset, which includes images captured at different magnification levels (40x, 100x, 200x, 400x), for both binary and multi classification of breast cancer subtypes. These low-weight hybrid ViT-CNN networks operate directly on input patches and convolution layers, to improve feature extraction and attention layers to train patches in all networks. This approach results in lower training time and fewer parameters while achieving accurate breast tumors classification. The proposed method is based on splitting the input image into patches and then focusing them on the area of cancerous lumps, providing a sequence of linear embedding of these patches as input. Second, we applied a convolution layer directly to the histopathology input patches, with the fewest possible modifications. Finally, we train patches in all transformer encoder layers to evaluate the performance of the classification of breast subtypes. The performance accuracies of our suggested models are 98.64% for VIT, 96.99% for CCT and 97.52% for MVIT. Moreover, the proposed models were compared with state-of-the-art models using the same dataset. Our study demonstrates how convolution and attention mechanisms can minimize computational training resources and decision time, to develop high- performing computer-aided analyses for breast cancer diagnosis. The source codes are accessible at https://github.com/abimouloud/ViT-CNN.



















Similar content being viewed by others
Data availability
The dataset analysed during the current study are available in: https://web.inf.ufpr.br/vri/databases/ breast-cancer-histopathological-database-breakhis/
References
Youlden DR et al (2012) The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol 36:237–248
Sohns C, Angic BC, Sossalla S, Konietschke F, Obenauer S (2010) Cad in full-field digital mammography—influence of reader experience and application of cad on interpretation of time. Clin Imaging 34:418–424
Saba T (2020) Recent advancement in cancer detection using machine learning: sys- tematic survey of decades, comparisons and challenges. J Infect Public Health 13:1274–1289
Nassif AB, Talib MA, Nasir Q, Afadar Y, Elgendy O (2022) Breast cancer detection using artificial intelligence techniques: a systematic literature review. Artif Intell Med 127:102276
Aggarwal R et al (2021) Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. NPJ digital medicine 4:65
Matsoukas C, Haslum JF, S¨oderberg M, Smith K (2021) Is it time to replace cnns with transformers for medical images? arXiv:2108.09038. Accessed 19 Jun 2023
Mohamed EA, Rashed EA, Gaber T, Karam O (2022) Deep learning model for fully automated breast cancer detection system from thermograms. PLoS ONE 17:e0262349
Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R (2019) A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 292:60–66
Henry EU, Emebob O, Omonhinmin CA (2022) Vision transformers in medical imaging: a review. arXiv:2211.10043. Accessed 19 Jun 2023
Dey RK, Das AK (2023) Modified term frequency-inverse document frequency based deep hybrid framework for sentiment analysis. Multimed Tools Appl 82:32967–32990
Dosovitskiy A. et al (2020) An image is worth 16x16 words: transformers for image recognition at scale. arXiv:2010.11929. Accessed 15 juin 2023
Zhu X, Cheng D, Zhang Z, Lin S, Dai J (2019) An empirical study of spatial attention mechanisms in deep networks. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp 6688–6697
Masood A, Naseem U, Kim J (2023) Multi-Level swin transformer enabled automatic segmentation and classification of breast metastases. In 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, pp 1–4. https://doi.org/10.1109/EMBC40787.2023.10340831
Dey RK, Das AK (2024) Neighbour adjusted dispersive flies optimization based deep hybrid sentiment analysis framework. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-17953-8
Hassani A. et al (2021) Escaping the big data paradigm with compact transformers. arXiv:2104.05704
Faheem M et al (2019) A multiobjective, lion mating optimization inspired routing protocol for wireless body area sensor network based healthcare applications. Sensors 19:5072
Alarood AA, Faheem M, Al-Khasawneh MA, Alzahrani AI, Alshdadi AA (2023) Secure medical image transmission using deep neural network in e-health applications. Healthcare Technol Lett 10:87–98
Iqbal S, Qureshi AN, Aurangzeb K, et al (2023) AMIAC: adaptive medical image analyzes and classification, a robust self-learning framework. Neural Comput & Applic. https://doi.org/10.1007/s00521-023-09209-1
Ali G, Dastgir A, Iqbal MW, Anwar M, Faheem M (2023) A hybrid convolutional neural network model for automatic diabetic retinopathy classification from fundus images. In IEEE Journal of Translational Engineering in Health and Medicine 11:341–350. https://doi.org/10.1109/JTEHM.2023.3282104
Wang P et al (2021) Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed Signal Process Control 65:102341
Albashish D, Al-Sayyed R, Abdullah A, Ryalat MH, Ahmad Almansour N (2021) Deep CNN Model based on VGG16 for breast cancer classification. In 2021 International Conference on Information Technology (ICIT), Amman, pp 805–810. https://doi.org/10.1109/ICIT52682.2021.9491631
Al-Jabbar M, Alshahrani M, Senan EM, Ahmed IA (2023) Multi-method diagnosis of histopathological images for early detection of breast cancer based on hybrid and deep learning. Mathematics 11:1429
Amin MS, Ahn H (2023) Fabnet: A features agglomeration-based convolutional neural network for multiscale breast cancer histopathology images classification. Cancers 15:1013
Hao Y et al (2022) Breast cancer histopathological images classification based on deep semantic features and gray level co-occurrence matrix. PLoS ONE 17:e0267955
Srikantamurthy MM, Rallabandi V, Dudekula DB, Natarajan S, Park J (2023) Classification of benign and malignant subtypes of breast cancer histopathology imaging using hybrid cnn-lstm based transfer learning. BMC Med Imaging 23:1–15
Mahmud MI, Mamun M, Abdelgawad A (2023) A deep analysis of transfer learning based breast cancer detection using histopathology images. In 2023 10th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, IEEE, pp 198–204, https://doi.org/10.1109/SPIN57001.2023.10117110
Abunasser BS, Al-Hiealy MRJ, Zaqout IS, Abu-Naser SS (2023) Con- volution neural network for breast cancer detection and classification using deep learning. Asian Pac J Cancer Preven: APJCP 24:531
Ayana G et al (2023) Vision-transformer-based transfer learning for mammogram classification. Diagnostics 13:178
He K et al (2023) Transformers in medical image analysis. Intell Med 3:59–78
Sriwastawa A, Arul Jothi JA (2024) Vision transformer and its variants for image classification in digital breast cancer histopathology: a comparative study. Multimed Tools Appl 83:39731–39753. https://doi.org/10.1007/s11042-023-16954-x
He L, Long LR, Antani S, Thoma GR (2012) Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed 107:538–556
ahmed IMb, Maalej R, Kherallah M (2023) MobileNet-Based model for histopathological breast cancer image classification. In: Abraham A, Hong TP, Kotecha K, Ma K, Manghirmalani Mishra P, Gandhi N (eds) Hybrid intelligent systems. HIS 2022. Lecture Notes in Networks and Systems, vol. 647. Springer, Cham. https://doi.org/10.1007/978-3-031-27409-1_58
Rulaningtyas R, Hyperastuty AS, Rahaju AS (2018) Histopathology grading identification of breast cancer based on texture classification using GLCM and neural network method. In Journal of Physics: Conference Series, vol. 1120, IOP Publishing, p 012050. https://doi.org/10.1088/1742-6596/1120/1/012050
He L, Long LR, Antani S, Thoma G (2010) Computer assisted diagnosis in histopathology. Sequenc Genome Anal: Methods Appl 15:271–287
Spanhol FA, Oliveira LS, Petitjean C, Heutte L (2015) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63:1455–1462
Breakhis - breast histopathology images dataset. https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/. Accessed 5 Jun 2023
Tummala S, Kim J, Kadry S (2022) Breast-net: multi-class classification of breast cancer from histopathological images using ensemble of swin transformers. Mathematics 10:4109
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30. https://doi.org/10.48550/arXiv.1706.03762
Mehta S, Rastegari M (2021) Mobilevit: light-weight, general-purpose, and mobile- friendly vision transformer. arXiv:2110.02178. Accessed 22 May 2023
Howard AG, et al (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv:1704.04861. Accessed 21 May 2023
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: inverted residuals and linear bottlenecks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520. https://doi.org/10.48550/arXiv.1801.04381
Howard A, Sandler M, Chu G, Chen L-C, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V, Le QV, Adam H (2019) Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)
Cheng Q, Li X, Zhu B, Shi Y, Xie B (2023) Drone detection method based on mobilevit and ca-panet. Electronics 12:223
Zou W, Xie K, Lin J (2023) Light-weight deep learning method for active jamming recognition based on improved mobilevit. Sonar & Navigation, IET Radar
Ahmed IA et al (2022) Eye tracking-based diagnosis and early detection of autism spectrum disorder using machine learning and deep learning techniques. Electronics 11:530
Saraswat D et al (2022) Explainable ai for healthcare 5.0: opportunities and challenges. IEEE Access 10:84486–84517
Chaddad A, Peng J, Xu J, Bouridane A (2023) Survey of explainable ai techniques in healthcare. Sensors 23:634
Wani NA, Kumar R, Bedi J, Rida I, et al (2024) Explainable AI-driven IoMT fusion: unravelling techniques, opportunities, and challenges with explainable AI in healthcare. Inf Fusion 102472. https://doi.org/10.1016/j.inffus.2024.102472
Acknowledgements
The authors extend their appreciation to the Deanship of Sci- entific Research at Northern Border University, Arar, KSA for funding this research work through the project number “ NBU-FFR-2024-2439-05.
Funding
This study was not funded. The authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
Authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
ABIMOULOUD, M.L., BENSID, K., Elleuch, M. et al. Vision transformer based convolutional neural network for breast cancer histopathological images classification. Multimed Tools Appl 83, 86833–86868 (2024). https://doi.org/10.1007/s11042-024-19667-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-024-19667-x