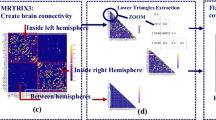
Abstract
Diagnosing Alzheimer’s disease (AD) in its prodromal stage is a significantly crucial area of research. Approximately 50% of individuals within the well-known Mild Cognitive Impairment (MCI) cohort are estimated to progress to AD, and the factors influencing conversion remain unknown. Gaining insights into the disease evolution can enhance support strategies and potentially slow down the pathology. Utilizing the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database, our objective is to construct a framework for distinguishing between Normal Controls (NC) and different stages of Alzheimer’s Disease (AD), encompassing Earlier Mild Cognitive Impairment (EMCI), Later Mild Cognitive Impairment (LMCI), and AD patients. In pursuit of this objective, we preprocessed Diffusion Tensor and Magnetic Resonance brain images from 237 subjects, generating corresponding brain connectivity maps. Notably, we introduce an innovative linearity assessment method that utilizes the Ordinary Least Squares (OLS) linear regression model to identify and select relevant features for classification. This approach effectively identifies features with strong linear relationships to the target variable. Our method’s superiority is demonstrated through a comparative analysis with the traditional SelectKBest approach. By integrating this feature selection strategy with a Logistic Regression model, our study achieves both efficient and highly accurate classification outcomes, highlighting the effectiveness of the proposed method. In a four-class classification scenario, the model attained an accuracy of \(66\% \pm 0.06\). In binary classification, the results were equally impressive, with an area under the curve of \(0.68\pm 0.10 \%\) for CN vs. EMCI discrimination, \(99\pm 0.02\% \)for distinguishing LMCI from adjacent classes CN and EMCI, and \(0.79\% \pm 0.08\) for discriminating AD from healthy subjects. Additionally, the calculation of Pearson’s correlation coefficient has been employed to identify cortical regions affected by changes, explore the nature of fiber disconnection propagation from one stage to another, and establish the traceability of the interference origin between stages. The summarized results reveal an apparent flow of white matter disruption from the right to the left hemisphere.







Similar content being viewed by others
References
Lock M (2013) The alzheimer conundrum. In: The Alzheimer Conundrum. Princeton University Press, ???
Jiji GW (2023) Biomarker to find neurodegenerative diseases using the structural changes in brain using computer vision. Multimed Tools Appl 82(22):34981–34993
Mabrouk B, BenHamida A, Drissi N, Bouzidi N (2023) Mhiri C Contribution of brain regions asymmetry scores combined with random forest classifier in the diagnosis of alzheimer’s disease in his earlier stage. J Med Biol Eng 43(1):74–82
Dickerson BC (2012) Wolk DA Mri cortical thickness biomarker predicts ad-like csf and cognitive decline in normal adults. Neurology 78(2):84–90
Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, Harvey DJ, Jack CR Jr, Weiner MW, Saykin AJ et al (2010) Longitudinal mri atrophy biomarkers: relationship to conversion in the adni cohort. Neurobiol Aging 31(8):1401–1418
Sabuncu MR, Desikan RS, Sepulcre J, Yeo BTT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA et al (2011) The dynamics of cortical and hippocampal atrophy in alzheimer disease. Arch Neurol 68(8):1040–1048
Jahanshahi AR, Naghdi Sadeh R, Khezerloo D (2023) Atrophy asymmetry in hippocampal subfields in patients with alzheimer’s disease and mild cognitive impairment. Exp Brain Res 241(2):495–504
Li A, Li F, Elahifasaee F, Liu M, Zhang L (2021) Hippocampal shape and asymmetry analysis by cascaded convolutional neural networks for alzheimer’s disease diagnosis. Brain Imaging Behav 15(5):2330–2339
Kim G-W, Kim B-C, Park KS, Jeong G-W (2020) A pilot study of brain morphometry following donepezil treatment in mild cognitive impairment: volume changes of cortical/subcortical regions and hippocampal subfields. Sci Rep 10(1):10912
Kim M, Kim SJ, Park JE, Yun J, Shim WH, Oh JS, Oh M, Roh JH, Seo SW, Oh SJ et al (2021) Combination of automated brain volumetry on mri and quantitative tau deposition on thk-5351 pet to support diagnosis of alzheimer’s disease. Sci Rep 11(1):10343
Uysal G, Ozturk M (2024) Comparative analysis of different brain regions using machine learning for prediction of emci and lmci stages of alzheimer’s disease. Multimed Tools Appl 83(7):21455–21470
Hirata Y, Matsuda H, Nemoto K, Ohnishi T, Hirao K, Yamashita F, Asada T, Iwabuchi S, Samejima H (2005) Voxel-based morphometry to discriminate early alzheimer’s disease from controls. Neurosci Lett 382(3):269–274
Busatto GF, Diniz BS, Zanetti MV (2008) Voxel-based morphometry in alzheimer’s disease. Expert review of neurotherapeutics 8(11):1691–1702
Schmitter D, Roche A, Maréchal B, Ribes D, Abdulkadir A, Bach-Cuadra M, Daducci A, Granziera C, Klöppel S, Maeder P et al (2015) An evaluation of volume-based morphometry for prediction of mild cognitive impairment and alzheimer’s disease. NeuroImage: Clinical 7:7–17
Aditya Shastry K, Sanjay H (2024) Artificial intelligence techniques for the effective diagnosis of alzheimer’s disease: a review. Multimed Tools Appl 83(13):40057–40092
Jazzar N, Mabrouk B, Douik A (2024) Cnl-resunet: A novel deep learning architecture for stroke image segmentation. In: 2024 IEEE 7th international conference on advanced technologies, signal and image processing (ATSIP), vol 1, pp 99–104. IEEE
Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, Xu M, Initiative ADN et al (2020) A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in alzheimer’s disease. Neuroimage 208:116459
Shahwar T, Zafar J, Almogren A, Zafar H, Rehman AU, Shafiq M, Hamam H (2022) Automated detection of alzheimer’s via hybrid classical quantum neural networks. Electronics 11(5):721
Tufail AB, Anwar N, Othman MTB, Ullah I, Khan RA, Ma Y-K, Adhikari D, Rehman AU, Shafiq M, Hamam H (2022) Early-stage alzheimer’s disease categorization using pet neuroimaging modality and convolutional neural networks in the 2d and 3d domains. Sensors 22(12):4609
Biswas R, Gini JR (2024) Multi-class classification of alzheimer’s disease detection from 3d mri image using ml techniques and its performance analysis. Multimed Tools Appl 83(11):33527–33554
Pei Z, Gou Y, Ma M, Guo M, Leng C, Chen Y, Li J (2022) Alzheimer’s disease diagnosis based on long-range dependency mechanism using convolutional neural network. Multimed Tools Appl, pp 1–16
Suchitra S, Krishnasamy L, Poovaraghan R (2024) A deep learning-based early alzheimer’s disease detection using magnetic resonance images. Multimed Tools Appl, pp 1–22
Ravi V, EA G, KP S et al (2024) Deep learning-based approach for multi-stage diagnosis of alzheimer’s disease. Multimed Tools Appl 83(6):16799–16822
Mabrouk B, Jazzar N, Sallemi L, Hamida AB (2024) A comparative study of pca and lda for dimensionality reduction in a 4-way classification framework. J App Mat Sci & Engg Res 8(1):1–6
Takahashi H, Ishii K, Kashiwagi N, Watanabe Y, Tanaka H, Murakami T, Tomiyama N (2017) Clinical application of apparent diffusion coefficient mapping in voxel-based morphometry in the diagnosis of alzheimer’s disease. Clin Radiol 72(2):108–115
Song S-K, Sun S-W, Ju W-K, Lin S-J, Cross AH, Neufeld AH (2003) Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage 20(3):1714–1722
Wang Q, Wang Y, Liu J, Sutphen CL, Cruchaga C, Blazey T, Gordon BA, Su Y, Chen C, Shimony JS,et al (2019) Quantification of white matter cellularity and damage in preclinical and early symptomatic alzheimer’s disease. NeuroImage: Clinical 22:101767
Bartzokis G (2011) Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging 32(8):1341–1371
Xue Y, Zhang Z, Wen C, Liu H, Wang S, Li J, Zhuge Q, Chen W, Ye Q (2019) Characterization of alzheimer’s disease using ultra-high b-values apparent diffusion coefficient and diffusion kurtosis imaging. Aging Dis 10(5):1026
Graña M, Termenon M, Savio A, Gonzalez-Pinto A, Echeveste J, Pérez J, Besga A (2011) Computer aided diagnosis system for alzheimer disease using brain diffusion tensor imaging features selected by pearson’s correlation. Neurosci Lett 502(3):225–229
La Rocca M, Amoroso N, Monaco A, Bellotti R, Tangaro S, Initiative ADN et al (2018) A novel approach to brain connectivity reveals early structural changes in alzheimer’s disease. Physiol Genomics 39(7):074005
Mabrouk B, Bouzidi N, Mhiri C, Hamida AB (2022) Combination of volumetric and topologic brain characteristics towards a diagnosis of alzheimer’s disease in his earlier stage. In: 2022 6th International conference on advanced technologies for signal and image processing (ATSIP), pp 1–4. IEEE
Mabrouk B, Hamida AB, Mabrouki N, Bouzidi N, Mhiri C (2024) A novel approach to perform linear discriminant analyses for a 4-way alzheimer’s disease diagnosis based on an integration of pearson’s correlation coefficients and empirical cumulative distribution function. Multimed Tools Appl, pp 1–17
Sporns O, Tononi G, Edelman GM (2000) Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb Cortex 10(2):127–141
Wu Z, Gao Y, Potter T, Benoit J, Shen J, Schulz PE, Zhang Y, Initiative ADN (2021) Interactions between aging and alzheimer’s disease on structural brain networks. Front Aging Neurosci 13:639795
Fischl B (2012) Freesurfer. Neuroimage 62(2):774–781
Amoroso N, Monaco A, Tangaro S (2017) Neuroimaging Initiative AD (2017) Topological measurements of dwi tractography for alzheimer’s disease detection. Comput Math Methods Med 1:5271627
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Duchesnay É (2011) Scikit-learn: Machine learning in python. J Mach Learn Res 12:2825–2830
Feng C, Wang H, Lu N, Chen T, He H, Lu Y, Tu X (2014) Log-transformation and its implications for data analysis. Shanghai Arch Psychiatry 26:105–109
Graham RL (1994) Concrete Mathematics: a Foundation for Computer Science. Pearson Education India, ???
Skipper Seabold, Josef Perktold (2010) Statsmodels: Econometric and Statistical Modeling with Python. In: Stéfan van der Walt, Jarrod Millman (eds.) Proceedings of the 9th python in science conference, pp 92–96. https://doi.org/10.25080/Majora-92bf1922-011
de Souza SV, Junqueira RG (2005) A procedure to assess linearity by ordinary least squares method. Anal Chim Acta 552(1–2):25–35
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay (2011) Scikit-learn: Machine learning in python. J Mach Learn Res 12:2825–2830
Liu S, Liu S, Cai W, Che H, Pujol S, Kikinis R, Feng D, Fulham MJ et al (2014) Multimodal neuroimaging feature learning for multiclass diagnosis of alzheimer’s disease. IEEE Trans Biomed Eng 62(4):1132–1140
Yao D, Calhoun VD, Fu Z, Du Y, Sui J (2018) An ensemble learning system for a 4-way classification of alzheimer’s disease and mild cognitive impairment. J Neurosci Methods 302:75–81
Lin W, Gao Q, Du M, Chen W, Tong T (2021) Multiclass diagnosis of stages of alzheimer’s disease using linear discriminant analysis scoring for multimodal data. Comput Biol Med 134:104478
Ruiz J, Mahmud M, Modasshir M, Shamim Kaiser M, Alzheimer’s Disease Neuroimaging Initiative f.t 3d densenet ensemble in 4-way classification of alzheimer’s disease. In: Brain informatics: 13th international conference, BI 2020, Padua, Italy, September 19, 2020, Proceedings 13, pp 85–96 (2020). Springer
Ghazal TM, Issa G (2022) Alzheimer disease detection empowered with transfer learning. Computers, Materials & Continua 70(3):5005–5019
Tang X, Liu J (2021) Comparing different algorithms for the course of alzheimer’s disease using machine learning. Ann Palliat Med 10(9):9715724–9719724
Park S, Kim SY et al (2018) A comparison between av45 and fdg-pet in alzheimer’s disease diagnosis. Int J Biome Imaging 2018:1247430. https://doi.org/10.1155/2018/1247430
Raghavaiah P, Varadarajan S (2021) A cad system design to diagnosize alzheimers disease from mri brain images using optimal deep neural network. Multimed Tools Appl 80(17):26411–26428
Ahmed HM, Elsharkawy ZF, Elkorany AS (2023) Alzheimer disease diagnosis for magnetic resonance brain images using deep learning neural networks. Multimed Tools Appl 82(12):17963–17977
Younes L, Albert M, Miller MI, Team BR,et al (2014) Inferring changepoint times of medial temporal lobe morphometric change in preclinical alzheimer’s disease. NeuroImage: Clinical 5:178–187
Zhu DC, Majumdar S, Korolev IO, Berger KL, Bozoki AC (2013) Alzheimer’s disease and amnestic mild cognitive impairment weaken connections within the default-mode network: a multi-modal imaging study. J Alzheimers Dis 34(4):969–984
Bi X-a, Xu Q, Luo X, Sun Q, Wang Z (2018) Analysis of progression toward alzheimer’s disease based on evolutionary weighted random support vector machine cluster. Front Neurol 12:716
Chang Y-T, Huang C-W, Chen N-C, Lin K-J, Huang S-H, Chang W-N, Hsu S-W, Hsu C-W, Chen H-H, Chang C-C (2016) Hippocampal amyloid burden with downstream fusiform gyrus atrophy correlate with face matching task scores in early stage alzheimer’s disease. Front Aging Neurosci 8:145
Tekin S, Mega MS, Masterman DM, Chow T, Garakian J, Vinters HV, Cummings JL (2001) Orbitofrontal and anterior cingulate cortex neurofibrillary tangle burden is associated with agitation in alzheimer disease. Ann Neurol 49(3):355–361
Rouw R, Scholte HS (2010) Neural basis of individual differences in synesthetic experiences. J Neurol 30(18):6205–6213
Van den Stock J, Tamietto M, Sorger B, Pichon S, Grézes J, de Gelder B (2011) Cortico-subcortical visual, somatosensory, and motor activations for perceiving dynamic whole-body emotional expressions with and without striate cortex (v1). Proc Natl Acad Sci 108(39):16188–16193
Lisowska A, Rekik I, AbbVie AA, Foundation ADD, Biotech A, Bio-Clinica I, Biogen Company B-MS, CereSpir I, Cogstate et al (2019) Joint pairing and structured mapping of convolutional brain morphological multiplexes for early dementia diagnosis. Brain Connectivity 9(1):22–36
Mejia-Vergara AJ, Karanjia R, Sadun AA (2021) Oct parameters of the optic nerve head and the retina as surrogate markers of brain volume in a normal population, a pilot study. J Neurol Sci 420:117213
den Haan J, Verbraak FD, Visser PJ, Bouwman FH (2017) Retinal thickness in alzheimer’s disease: a systematic review and meta-analysis. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring 6:162–170
Neufang S, Akhrif A, Riedl V, Förstl H, Kurz A, Zimmer C, Sorg C, Wohlschläger AM (2011) Disconnection of frontal and parietal areas contributes to impaired attention in very early alzheimer’s disease. J Alzheimers Dis 25(2):309–321
Grambaite R, Selnes P, Reinvang I, Aarsland D, Hessen E, Gjerstad L, Fladby T (2011) Executive dysfunction in mild cognitive impairment is associated with changes in frontal and cingulate white matter tracts. J Alzheimers Dis 27(2):453–462
Parker TD, Slattery CF, Zhang J, Nicholas JM, Paterson RW, Foulkes AJ, Malone IB, Thomas DL, Modat M, Cash DM et al (2018) Cortical microstructure in young onset alzheimer’s disease using neurite orientation dispersion and density imaging. Hum Brain Mapp 39(7):3005–3017
McDonald C, McEvoy L, Gharapetian L, Fennema-Notestine C, Hagler D, Holland D, Koyama A, Brewer J, Dale A et al (2009) Regional rates of neocortical atrophy from normal aging to early alzheimer disease. Neurology 73(6):457–465
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through a large group Research Project under grant number RGP2/390/45.
Author information
Authors and Affiliations
Contributions
BM and ABH were responsible for the conception and study design. Statistical analysis was conducted by BM , and NM, while the interpretation of results involved BM, NM, NB, and LS. BM managed the dataset acquisition and preprocessing, as well as the development and implementation of the coding. BM, and NM drafted the manuscript and revised it critically for important intellectual content. All authors approved the final version to be published and agreed to be accountable for the integrity and accuracy of all aspects of the work.
Corresponding author
Ethics declarations
Competing of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at:(http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf) .
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mabrouk, B., Bouattour, N., Mabrouki, N. et al. A novel approach to enhance feature selection using linearity assessment with ordinary least squares regression for Alzheimer’s Disease stage classification. Multimed Tools Appl 83, 86059–86078 (2024). https://doi.org/10.1007/s11042-024-20254-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-024-20254-3