Abstract
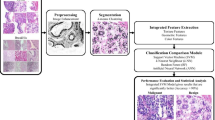
Oral squamous cell carcinoma (OSCC) diagnosis through computer vision approach is newly introduced technique in the modern diagnostic era. Mitotic cell count from related tissue histopathological images signifies the proliferative marker of cancer cell has been recognized as an essential phenomenon in diagnosis. This paper aims at developing an automated technique for accomplishing the task of mitotic cell count from related histopathological images. In this regard, a new machine learning based methodology incorporating random forest tree classifier learns over four entropy measures, fractal dimension, and seven Hu’s moments based descriptors have been introduced. The performance validation summarizes that proposed methodology can detect mitotic cell efficiently from histopathological images of OSCC with 89% precision, 95% recall or sensitivity, 97.35% specificity, 96.92% accuracy, 96.45% AUC and 92% F-score measure.










Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
ABPI. (2015). Cell division and cancer. http://www.abpischools.org.uk/page/modules/celldiv_cancer/cancer4.cfm. Accessed 23 Jan 2015.
Aloraidi, N. A., Sirinukunwattana, K., Khan, A. M., & Rajpoot, N. M. (2014). On generating cell exemplars for detection of mitotic cells in breast cancer histopathology images. In Proceedings of the 36th annual international conference of the IEEE engineering in medicine and biology society (EMBC)(pp. 3370–3373). doi:10.1109/EMBC.2014.6944345.
Anneroth, G., Batskis, J., & Luna, M. (1987). Review of the literature and a recommended system of malignancy grading in oral squamous cell carcinomas. European Journal of Oral Sciences, 95(3), 229–249.
Breiman, L. (2001). Random forests. Machine learning, 45(1), 5–32.
Chen, W., Wang, Y., Cao, G., Chen, G., & Gu, Q. (2014). A random forest model based classification scheme for neonatal amplitude-integrated EEG. BioMedical Engineering OnLine,. doi:10.1186/1475-925X-13-S2-S4.
Cireşan, D. C., Giusti, A., Gambardella, L. M., & Schmidhuber, J. (2013). Mitosis detection in breast cancer histology images with deep neural networks. In Proceedings of the medical image computing and computer-assisted intervention(MICCAI 2013) (pp. 411–418). Springer.
Das, D., Ghosh, M., Chakraborty, C., Pal, M., & Maity, A. K. (2010). Invariant moment based feature analysis for abnormal erythrocyte recognition. InProceedings of the IEEE international conference on systems in medicine and biology (ICSMB 2010) (pp. 242–247).
Das, D. K., Ghosh, M., Pal, M., Maiti, A. K., & Chakraborty, C. (2013). Machine learning approach for automated screening of malaria parasite using light microscopic images. Micron, 45, 97–106.
Das, D. K., Koley, S., Chakraborty, C., & Maiti, A. K. (2014). Automated segmentation of Mitotic Cells for in vitro histological evaluation of oral squamous cell carcinoma. In Proceedings of the IEEE international symposium on signal processing and information technology (ISSPIT) (pp. 000354–000357).
Gallardo, G. M., Yang, F., Ianzini, F., Mackey, M., & Sonka, M. (2004). Mitotic cell recognition with hidden Markov models. In Proceedings SPIE 5367, Medical Imaging 2004: Visualization, Image-Guided Procedures, and Display, 661, San Diago, CA. doi:10.1117/12.535778.
Ghosh, M., Das, D., & Chakraborty, C. (2010). Entropy-based divergence for leukocyte image segmentation. In Proceedings of the IEEE international conference on systems in medicine and biology (ICSMB2010) (pp. 409–413).
Giusti, A., Caccia, C., Ciresan, D. C., Schmidhuber, J., & Gambardella, L. M. (2014). A comparison of algorithms and humans for mitosis detection. In Proceedings of the IEEE 11th international symposium on biomedical imaging (ISBI)(pp. 1360–1363). doi:10.1109/ISBI.2014.6868130.
Gonzalez, R. C. (2009). Digital image processing. Bengaluru: Pearson Education India.
Hu, M.-K. (1962). Visual pattern recognition by moment invariants. IRE Transactions on Information Theory, 8(2), 179–187.
Irshad, H. (2013). Automated mitosis detection in histopathology using morphological and multi-channel statistics features. Journal of Pathology Informatics, 4(1), 10.
Khan, A. M., Eldaly, H., & Rajpoot, N. M. (2013). A gamma-gaussian mixture model for detection of mitotic cells in breast cancer histopathology images. Journal of Pathology Informatics, 4(1), 11.
Khan, A. M., Rajpoot, N., Treanor, D., & Magee, D. (2014). A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Transactions on Biomedical Engineering, 61(6), 1729–1738.
Kurt, B., Nabiyev, V. V., & Turhan, K. (2014). A novel automatic suspicious mass regions identification using Havrda & Charvat entropy and Otsu’s N thresholding. Computer Methods and Programs in Biomedicine, 114(3), 349–360.
Lu, C., Ji, M., Ma, Z., & Mandal, M. (2015). Automated image analysis of nuclear atypia in high-power field histopathological image. Journal of Microscopy, 258(3), 233–240.
Lu, C., & Mandal, M. (2014). Toward automatic mitotic cell detection and segmentation in multispectral histopathological images. IEEE Journal of Biomedical and Health Informatics, 18(2), 594–605.
Macenko, M., Niethammer, M., Marron, J., Borland, D., Woosley, J., Guan, X., Schmitt, C., Thomas, N. (2009). A method for normalizing histology slides for quantitative analysis. In:Proceedings of the sixth IEEE international symposium on biomedical imaging (ISBI) (pp. 1107–1110).
Malon, C. D., & Cosatto, E. (2013). Classification of mitotic figures with convolutional neural networks and seeded blob features. Journal of Pathology Informatics, 4(1), 9.
Mandelbrot, B. B. (1983). The fractal geometry of nature (Vol. 173). London: Macmillan.
Nateghi, R., Danyali, H., Helfroush, M. S., & Tashk, A. (2014a). Mitosis detection from breast cancer histology slide images using particle swarm optimization and support vector machine. International Journal of Sciences: Basic and Applied Research (IJSBAR), 16(1), 164–177.
Nateghi, R., Danyali, H., SadeghHelfroush, M., & Pour, F. P. (2014b). Automatic detection of mitosis cell in breast cancer histopathology images using a genetic algorithm. In Proceedings of the IEEE 21th Iranian conference on biomedical engineering (ICBME) (pp. 1–6).
Paul, A., & Mukherjee, D. P. (2014). Enhanced random forest for mitosis detection. In Proceedings of the 2014 Indian conference on computer vision graphics and image processing, Vol. 14 (pp. 85). Bangalore, India. doi:10.1145/2683483.2683569.
Pharwaha, A. P. S., & Singh, B. (2009). Shannon and non-shannon measures of entropy for statistical texture feature extraction in digitized mammograms. In Proceedings of the world congress on engineering and computer science, Vol. 2(pp. 20–22).
Powers, D. M. (2011). Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. http://hdl.handle.net/2328/27165. Accessed 20 July 2014.
Ranking, W. H. (2014). Oral Cancer. http://www.worldlifeexpectancy.com/cause-of-death/oral-cancer/by-country/. Accessed Oct 2014.
Sarkar, N., & Chaudhuri, B. (1994). An efficient differential box-counting approach to compute fractal dimension of image. IEEE Transactions on Systems, Man and Cybernetics, 24(1), 115–120.
Şerbănescu, M. -S. (2013). Semi-automated mitosis detection in histopathological images of breast. Annals Computer Science Series, 11(1), 66–70
Sommer, C., Fiaschi, L., Hamprecht, F. A., & Gerlich, D. (2012). Learning-based mitotic cell detection in histopathological images. In Proceedings of the IEEE 21st international conference on pattern recognition (ICPR) (pp. 2306–2309).
Tashk, A., Helfroush, M. S., Danyali, H., & Akbarzadeh-jahromi, M. (2015). Automatic detection of breast cancer mitotic cells based on the combination of textural, statistical and innovative mathematical features. Applied Mathematical Modelling, 39(20), 6165–6182.
Tashk, A., Helfroush, M. S., Danyali, H., & Akbarzadeh, M. (2014). A novel CAD system for mitosis detection using histopathology slide images. Journal of Medical Signals and Sensors, 4(2), 139–149.
Tek, F. B. (2013). Mitosis detection using generic features and an ensemble of cascade adaboosts. Journal of Pathology Informatics, 4(1), 12. doi:10.4103/2153-3539.112697.
Veta, M., van Diest, P. J., & Pluim, J. P. W. (2013). Detecting mitotic figures in breast cancer histopathology images. In Proceedings of the SPIE medical imaging (Digital Pathology), 8676, pp. 867607–867607. doi:10.1117/12.2006626.
Vink, J. P., Van Leeuwen, M. B., Van Deurzen, C. H. M., & De Haan, G. (2013). Efficient nucleus detector in histopathology images. Journal of Microscopy, 249(2), 124–135.
Wan, T., Liu, L., Chen, J., & Qin, Z. (2014). Wavelet-based statistical features for distinguishing mitotic and non-mitotic cells in breast cancer histopathology. In Proceedings of the IEEE international conference on image processing (ICIP) (pp. 2290–2294). doi:10.1109/ICIP.2014.7025464.
Wang, H., Cruz-Roa, A., Basavanhally, A., Gilmore, H., Shih, N., & Feldman, M., et al. (2014). Cascaded ensemble of convolutional neural networks and handcrafted features for mitosis detection. In Proceedings of the SPIE medical imaging, Vol. 9041, pp. 90410B–90410B. doi:10.1117/12.2043902.
Acknowledgements
The D.K.D. would like to acknowledge Council of Scientific and Industrial Research (CSIR), India for providing financial support to carry out this research under CSIR-SRF scheme (09/81(1203)/2013/EMR-I date. 16.03.2013). C.C., P.M., S.C., A.K.M. and S.B. acknowledge the support provided by Ministry of Human Resource Development, Govt. of India under the Grant Ref. No. 4-23/2014 T.S.I. Date: 14-02-2014. Authors are thankful to the reviewers for their valuable suggestions to improve the quality of this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Das, D.K., Mitra, P., Chakraborty, C. et al. Computational approach for mitotic cell detection and its application in oral squamous cell carcinoma. Multidim Syst Sign Process 28, 1031–1050 (2017). https://doi.org/10.1007/s11045-017-0488-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11045-017-0488-6