Abstract
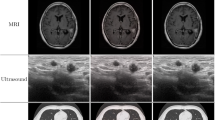
In this paper, we present a novel statistical approach to medical image segmentation. This approach is based on finite mixture models with spatial smoothness constrains. The main advantages of the proposed approach can be summarized as follows. Firstly, the proposed model is based on inverted Dirichlet mixture models, which have demonstrated better performance in modeling positive data (e.g., images) than Gaussian mixture models. Secondly, we integrate spatial relationships between pixels with the inverted Dirichlet mixture model, which makes it more robust against noise and image contrast levels. Finally, we develop a variational Bayes method to learn the proposed model, such that the model parameters and model complexity (i.e., the number of mixture components) can be estimated simultaneously in a unified framework. The performance of the proposed approach in medical image segmentation is compared with some state-of-the-art segmentation approaches through various numerical experiments on both simulated and real medical images.









Similar content being viewed by others
References
Ahmed MN, Yamany SM, Mohamed N, Farag AA, Moriarty T (2002) A modified fuzzy c-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans Med Imaging 21(3):193–199
Alfò M, Nieddu L, Vicari D (2008) A finite mixture model for image segmentation. Stat Comput 18(2):137–150
Ashburner J, Friston KJ (2005) Unified segmentation. NeuroImage 26(3):839–851
Attias H (1999) A variational Bayes framework for graphical models. In: Proceedings of of Advances in Neural Information Processing Systems (NIPS), pp 209–215
Bdiri T, Bouguila N (2012) Positive vectors clustering using inverted Dirichlet finite mixture models. Expert Syst Appl 39(2):1869–1882
Bdiri T, Bouguila N (2013) Bayesian learning of inverted Dirichlet mixtures for SVM kernels generation. Neural Comput Appl 23(5):1443–1458
Bishop CM (2006) Pattern recognition and machine learning. Springer, Berlin
Blei DM, Jordan MI (2005) Variational inference for Dirichlet process mixtures. Bayesian Anal 1:121–144
Blekas K, Likas A, Galatsanos NP, Lagaris IE (2005) A spatially constrained mixture model for image segmentation. IEEE Trans Neural Netw 16(2):494–498
Celeux G, Forbes F, Peyrard N (2003) EM procedures using mean field-like approximations for Markov model-based image segmentation. Pattern Recogn 36(1):131–144
Chatzis SP, Varvarigou TA (2008) A fuzzy clustering approach toward hidden Markov random field models for enhanced spatially constrained image segmentation. IEEE Trans Fuzzy Syst 16(5):1351–1361
Choi HS, Haynor DR, Kim Y (1991) Partial volume tissue classification of multichannel magnetic resonance images—a mixel model. IEEE Trans Med Imaging 10(3):395–407
Cocosco CA, Kollokian V, Kwan RKS, Pike GB, Evans AC (1997) BrainWeb: Online interface to a 3D MRI simulated brain database. NeuroImage 5:425
Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC (1998) Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging 17(3):463–468
Comaniciu D, Meer P (2002) Mean shift: a robust approach toward feature space analysis. IEEE Trans Pattern Anal Mach Intell 24(5):603–619
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Fan W, Bouguila N, Ziou D (2012) Variational learning for finite Dirichlet mixture models and applications. IEEE Trans Neural Netw Learn Syst 23(5):762–774
Forbes F, Peyrard N (2003) Hidden Markov random field model selection criteria based on mean field-like approximations. IEEE Trans Pattern Anal Mach Intell 25(9):1089–1101
Hong C, Yu J, Tao D, Wang M (2015a) Image-based three-dimensional human pose recovery by multiview locality-sensitive sparse retrieval. IEEE Trans Ind Electron 62(6):3742–3751
Hong C, Yu J, Wan J, Tao D, Wang M (2015b) Multimodal deep autoencoder for human pose recovery. IEEE Trans Image Process 24(12):5659–5670
Hong C, Yu J, You J, Chen X, Tao D (2015c) Multi-view ensemble manifold regularization for 3D object recognition. Inf Sci 320:395–405
Kwan RKS, Evans AC, Pike GB (1996) An extensible MRI simulator for post-processing evaluation. In: Proceedings of the 4th international conference on visualization in biomedical computing (VBC’96), pp 135–140
Kwan RKS, Evans AC, Pike GB (1999) MRI simulation-based evaluation of image-processing and classification methods. IEEE Trans Med Imaging 18(11):1085–1097
Li C, Kao CY, Gore JC, Ding Z (2008) Minimization of region-scalable fitting energy for image segmentation. IEEE Trans Image Process 17(10):1940–1949
Luque-Baena RM, Ortiz-de Lazcano-Lobato JM, López-Rubio E, Palomo EJ (2013) A competitive neural network for multiple object tracking in video sequence analysis. Neural Process Lett 37(1):47–67
McLachlan G, Peel D (2000) Finite mixture models. Wiley, New York
Mentzelopoulos M, Psarrou A, Angelopoulou A, García-Rodríguez J (2013) Active foreground region extraction and tracking for sports video annotation. Neural Process Lett 37(1):33–46
Nasios N, Bors AG (2006) Variational learning for Gaussian mixture models. IEEE Trans Syst Man Cybern B Cybern 36(4):849–862
Nguyen N, Wu QMJ, Ahuja S (2010) An extension of the standard mixture model for image segmentation. IEEE Trans Neural Netw 21(8):1326–1338
Nguyen TM, Wu QMJ (2012) Robust student’s-t mixture model with spatial constraints and its application in medical image segmentation. IEEE Trans Med Imaging 31(1):103–116
Nikou C, Galatsanos NP, Likas AC (2007) A class-adaptive spatially variant mixture model for image segmentation. IEEE Trans Image Process 16(4):1121–1130
Nikou C, Likas AC, Galatsanos NP (2010) A bayesian framework for image segmentation with spatially varying mixtures. IEEE Trans Image Process 19(9):2278–2289
Pham DL (2001) Spatial models for fuzzy clustering. Comput Vis Image Underst 84(2):285–297
Robert C, Casella G (1999) Monte Carlo statistical methods. Springer, Berlin
Salembier P, Garrido L (2000) Binary partition tree as an efficient representation for image processing, segmentation, and information retrieval. IEEE Trans Image Process 9(4):561–576
Sanjay GS, Hebert TJ (1998) Bayesian pixel classification using spatially variant finite mixtures and the generalized EM algorithm. IEEE Trans Image Process 7(7):1014–1028
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905
Tiao GG, Cuttman I (1965) The inverted Dirichlet distribution with applications. J Am Stat Assoc 60(311):793–805
Titterington DM, Smith AFM, Makov UE (1985) Statistical analysis of finite mixture distributions. Wiley, Hoboken
Yu J, Tao D, Wang M (2012) Adaptive hypergraph learning and its application in image classification. IEEE Trans Image Process 21(7):3262–3272
Yu J, Tao D, Li J, Cheng J (2014) Semantic preserving distance metric learning and applications. Inf Sci 281:674–686
Zhang Y, Brady M, Smith S (2001) Segmentation of brain mr images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20(1):45–57
Acknowledgements
The completion of this work was supported by the National Natural Science Foundation of China (61502183, 61673186), and the Scientific Research Funds of Huaqiao University (600005- Z15Y0016).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, W., Hu, C., Du, J. et al. A Novel Model-Based Approach for Medical Image Segmentation Using Spatially Constrained Inverted Dirichlet Mixture Models. Neural Process Lett 47, 619–639 (2018). https://doi.org/10.1007/s11063-017-9672-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-017-9672-9