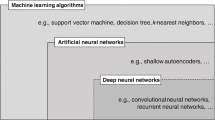
Abstract
The concept of deep learning originates from artificial neural networks which has become a very popular research area during the past few decades. There are two main reasons for for the wide acceptance of deep learning. First one being the overfitting problem has been partially resolved with the advent of big data analytics techniques. The second point for wide acceptance of deep learning is that deep neural networks undergo pre-training procedure before unsupervised learning, which assigns some initial values to the network. This article describes the all deep learning techniques and their experimental analysis with advantage and disadvantages. This review highlights the till date progress of the six deep learning techniques namely, autoencoder, restricted Boltzmann machine, deep belief network, recurrent neural network, convolutional neural network, and generative adversarial network with practical variant case studies. A wide discourse has been taken into consideration for the survey in the article. It concludes try to reflect some of the most fundamental and recent applications in the medical health-care system, and also identify some of the challenges and opportunities of the deep learning techniques.








Similar content being viewed by others
Change history
06 July 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11063-021-10527-5
References
Li D (2014) A tutorial survey of architectures, algorithms, and applications for deep learning. APSIPA Trans Signal Inf Process 3:e2
Bengio Y (2009) Learning deep architectures for AI. Found Trends Mach Learn 2(1):1–127
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Hinton GE (2006) Reducing the dimensionality of data with neural networks. Science (80) 313(5786):504–507
Bengio Y, Lamblin P, Popovici D, Larochelle H (2007) Greedy layer-wise training of deep networks. Adv Neural Inf Process Syst 19(1):153
Silver D et al (2016) Mastering the game of Go with deep neural networks and tree search. Nature 529(7587):484–489
Hinton RSZ, Geoffrey E (1994) Autoencoders, minimum description length and Helmholtz free energy. Adv Neural Inf Process Syst 3(3):219
Hinton GE, Salakhutdinov RR (2006) Reducing the dimensionality of data with neural networks. Science (80) 313(5786):504–507
Meyer D (2015) Introduction to autoencoders
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828
Dietterich T (1995) Overfitting and undercomputing in machine learning. ACM Comput Surv 27(3):326–327
Palm RB (2012) Prediction as a candidate for learning deep hierarchical models of data. Technical University of Denmark, 5
Pérez-Cruz F (2008) Kullback-Leibler divergence estimation of continuous distributions. In: Information theory, 2008. ISIT 2008. IEEE international symposium on. IEEE, pp 1666–1670
Widrow B, Lehr MA (1990) 30 Years of adaptive neural networks: perceptron, madaline, and backpropagation. Proc IEEE 78(9):1415–1442
Vincent P, Larochelle H, Bengio Y, Manzagol P-A (2008) Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th international conference on machine learning (ICML 2008), pp 1096–1103
Rifai S, Muller X (2011) Contractive auto-encoders: explicit invariance during feature extraction. ICML 85(1):833–840
Makhzani A, Frey B (2013) K-sparse autoencoders. arXiv preprint arXiv:1312.5663
Sun M, Zhang X, Van Hamme H, Zheng TF (2016) Unseen noise estimation using separable deep auto encoder for speech enhancement. IEEE ACM Trans Speech Lang Process 24(1):93–104
Ackley DH, Hinton GE, Sejnowski TJ (1985) A learning algorithm for Boltzmann machines. Cogn Sci 9:147–169
Hinton G (2010) A practical guide to training restricted Boltzmann machines a practical guide to training restricted Boltzmann machines. Computer (Long. Beach. Calif) 9(3):1
Hinton GE (2002) Training products of experts by minimizing contrastive divergence. Neural Comput 14(8):1771–1800
Fischer A, Igel C (2012) An introduction to restricted Boltzmann machines. Lect Notes Comput Sci Prog Pattern Recogn Image Anal Comput Vis Appl 7441:14–36
Bengio Y, Delalleau O (2009) Justifying and generalizing contrastive divergence. Neural Comput 21(6):1601–1621
Sutskever I, Tieleman T (2010) On the convergence properties of contrastive divergence. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp 789–795
Fischer A, Igel C (2011) Training RBMs based on the signs of the CD approximation of the log-likelihood derivatives. In: ESANN
Mnih V, Larochelle H, Hinton GE (2012) Conditional restricted boltzmann machines for structured output prediction. arXiv preprint arXiv:1202.3748
Larochelle H, Bengio Y (2008) Classification using discriminative restricted Boltzmann machines. In: ICML, pp 536–543
Hinton GE (2012) A practical guide to training restricted boltzmann machines BT. In: Montavon G, Orr GB, Mller K-R (eds) Neural Networks: tricks of the trade, Second edn. Springer, Berlin, pp 599–619
Tang Y, Salakhutdinov R, Hinton G (2012) Robust Boltzmann machines for recognition and denoising. In: Proceedings of the IEEE conference on computer vision and pattern Recognition, pp 2264–2271
Keyvanrad MA, Homayounpour MM A (2014, September) brief survey on deep belief networks and introducing a new object oriented MATLAB toolbox (DeeBNet). In: CoRR, vol. abs/1408.3, pp 0–25
Deng L (2013) Three classes of deep learning architectures and their applications: a tutorial survey. Microsoft Research, Washington
Arnold L, Rebecchi S, Chevallier S, Paugam-Moisy H (2011) An introduction to deep learning. In: European Symposium on Artificial Neural Networks (ESANN), Apr 2011, Bruges, Belgium. Proceedings of the European Symposium on Artificial Neural Networks
Plis SM et al (2014) Deep learning for neuroimaging: a validation study. Front Neurosci 8:229
Hinton GE, Dayan P, Frey BJ, Neal RM (1995) The “wake-sleep” algorithm for unsupervised neural networks. Science (80) 268(5214):1158–1161
Tang A, Lu K, Wang Y, Huang J, Li H (2015) A real-time hand posture recognition system using deep neural networks. ACM Trans Intell Syst Technol 6(2):21:1–21:23
Salakhutdinov R, Hinton G (2008) Using deep belief nets to learn covariance kernels for Gaussian processes. Adv Neural Inf Process Syst (20) 20:1–8
Nair V, Hinton GE (2009) 3D object recognition with deep belief nets. Adv Neural Inf Process Syst 21:1339–1347
Kuremoto T, Kimura S, Kobayashi K, Obayashi M (2014) Time series forecasting using a deep belief network with restricted Boltzmann machines. Neurocomputing 137:47–56
Liao B, Xu J, Lv J, Zhou S (2015) An image retrieval method for binary images based on DBN and Softmax classifier. IETE Tech Rev 32(4):294–303
Abdel-Zaher AM, Eldeib AM (2016) Breast cancer classification using deep belief networks. Expert Syst Appl 46:139–144
Qin M, Li Z, Du Z (2017) Red tide time series forecasting by combining ARIMA and deep belief network. Knowl Based Syst 125:39–52
Mandic DP, Chambers JA (2001) Recurrent neural networks for prediction: learning algorithms, architectures and stability. Wiley, New York, pp 171–198
Bengio Y, Simard P, Frasconi P (1994) Learning long-term dependencies with gradient descent is difficult. IEEE Trans Neural Netw 5(2):157–166
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Sak H, Senior A, Beaufays F (2014) Long short-term memory recurrent neural network architectures for large scale acoustic modeling. In: Fifteenth annual conference of the international speech communication association
Chung J, Gulcehre C, Cho K, Bengio Y (2015, June) Gated feedback recurrent neural networks. In: International conference on machine learning, pp 2067–2075
Yildirim O (2018) A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput Biol Med 96:189–202
Jin B, Che C, Liu Z, Zhang S, Yin X, Wei X (2018) Predicting the risk of heart failure with EHR sequential data modeling. IEEE Access 6:9256–9261
Zhao A, Qi L, Li J, Dong J, Yu H (2018, April) LSTM for diagnosis of neurodegenerative diseases using gait data. In: Ninth international conference on graphic and image processing (ICGIP 2017), international society for optics and photonics, vol 10615, p 106155B
Hubel DH, Wiesel TN (1968) Receptive fields and functional architecture of monkey striate cortex. J Physiol 195:215–243
Fukushima K (1980) Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern 36(4):193–202
Lang KJ, Waibel AH, Hinton GE (1990) A time-delay neural network architecture for isolated word recognition. Neural Netw 3(1):23–43
Le Cun Jackel LD, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Le Cun B, Denker J, Henderson D (1990) Handwritten digit recognition with a back-propagation network. Adv Neural Inf Process Syst 2:396–404
Hecht-Nielsen R (1989) Theory of the backpropagation neural network. Proc Int Jt Conf Neural Netw 1:593–605
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2323
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv. Neural Inf Process Syst 25:1–9
Nair V, Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th international conference on machine learning, no. 3, pp 807–814
Boureau Y-L, Ponce J, LeCun Y (2010) A theoretical analysis of feature pooling in visual recognition. In: ICML, pp 111–118
Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M (2014) Striving for simplicity: the all convolutional net. arXiv preprint arXiv:1412.6806
Eigen D, Rolfe J, Fergus R, LeCun Y (2013) Understanding deep architectures using a recursive convolutional network. arXiv preprint arXiv:1312.1847
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In: European conference on computer vision. Springer, Cham, pp 818–833
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Ponti MA, Ribeiro LS, Nazare TS, Bui T, Collomosse J (2018) Everything you wanted to know about deep learning for computer vision but were afraid to ask. In: 2017 30th SIBGRAPI conference on graphics, patterns and images tutorials (SIBGRAPI-T)
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Bengio Y (2014) Generative adversarial nets. In: Advances in neural information processing systems, pp 2672–2680
Radford A, Metz L, Chintala S (2015) Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434
Huang X, Li Y, Poursaeed O, Hopcroft J, Belongie S (2017) Stacked generative adversarial networks. IEEE Conf Comput Vis Pattern Recogn 2:4
Goodfellow I (2016) NIPS 2016 tutorial: generative adversarial networks. arXiv preprint arXiv:1701.00160
Zhai S, Cheng Y, Feris R, Zhang Z (2016) Generative adversarial networks as variational training of energy based models. arXiv preprint arXiv:1611.01799
Reed S, Akata Z, Yan X, Logeswaran L, Schiele B, Lee H (2016) Generative adversarial text to image synthesis. arXiv preprint arXiv:1605.05396
Kamnitsas K, Baumgartner C, Ledig C, Newcombe V, Simpson J, Kane A, Glocker B (2017, June) Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: International conference on information processing in medical imaging. Springer, Cham, pp 597–609
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. arXiv preprint
Al Rahhal MM et al (2016) Deep learning approach for active classification of electrocardiogram signals. Inf Sci 345:340–354
Kooi T et al (2017) Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 35:303–312
Ching T et al (2017, May) Opportunities and obstacles for deep learning in biology and medicine. bioRxiv
Najafabadi MM, Villanustre F, Khoshgoftaar TM, Seliya N, Wald R, Muharemagic E (2015) Deep learning applications and challenges in big data analytics. J Big Data 2(1):1
Suthaharan S (2014) Big data classification. ACM Sigmetrics Perform Eval Rev 41(4):70–73
Glorot X, Bordes A, Bengio Y (2011) Domain adaptation for large-scale sentiment classification: a deep learning approach. In: Proceedings of the 28th international conference on machine learning, no. 1, pp 513–520
Chen X-W, Lin X (2014) Big data deep learning: challenges and perspectives. IEEE Access 2:514–525
Brosch T, Tam R, Alzheimers Disease Neuroimaging Initiative (2013, September) Manifold learning of brain MRIs by deep learning. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 633–640
Suk HI, Shen D (2013, September) Deep learning-based feature representation for AD/MCI classification. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 583–590
Plis SM, Hjelm DR, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, Calhoun VD (2014) Deep learning for neuroimaging: a validation study. Front Neurosci 8:229
Yang D, Zhang S, Yan Z, Tan C, Li K, Metaxas D (2015, April) Automated anatomical landmark detection ondistal femur surface using convolutional neural network. In: 2015 IEEE 12th international symposium on biomedical imaging (ISBI), pp 17–21
Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D (2015, October) 3D deep learning for efficient and robust landmark detection in volumetric data. In International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 565–572
Ghesu FC, Georgescu B, Mansi T, Neumann D, Hornegger J, Comaniciu D (2016, October) An artificial agent for anatomical landmark detection in medical images. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 229–237
Baumgartner CF, Kamnitsas K, Matthew J, Smith S, Kainz B, Rueckert D (2016, October) Real-time standard scan plane detection and localisation in fetal ultrasound using fully convolutional neural networks. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 203–211
Chen H, Dou Q, Ni D, Cheng JZ, Qin J, Li S, Heng PA (2015, October) Automatic fetal ultrasound standard plane detection using knowledge transferred recurrent neural networks. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 507–514
Chen J, Yang L, Zhang Y, Alber M, Chen DZ (2016) Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. In: Advances in neural information processing systems, pp 3036–3044
Xie Y, Zhang Z, Sapkota M, Yang L (2016, October) Spatial clockwork recurrent neural network for muscle perimysium segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 185–193
Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D (2013, September) Unsupervised deep feature learning for deformable registration of MR brain images. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 649–656
Cheng X, Zhang L, Zheng Y (2018) Deep similarity learning for multimodal medical images. Comput Methods Biomech Biomed Eng Imaging Vis 6(3):248–252
Krebs J, Mansi T, Delingette H, Zhang L, Ghesu FC, Miao S, Kamen A (2017, September) Robust non-rigid registration through agent-based action learning. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 344–352
Simonovsky M, Gutirrez-Becker B, Mateus D, Navab N, Komodakis N (2016) A deep metric for multimodal registration. In: Proceedings of the medical image computing and computer-assisted intervention. Lecture notes in computer science, 9902, pp 10–18
Greenspan H, van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35(5):1153–1159
Nie D, Zhang H, Adeli E, Liu L, Shen D (2016) 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients BT-medical image computing and computer-assisted intervention. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds) Proceeding of the MICCAI 2016: 19th international conference, Athens, Greece, 17–21 October 2016. Springer International Publishing, Cham, pp 212–220
Sun W, Tseng TB, Zhang J, Qian W (2017) Computerized medical imaging and graphics enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graph 57:4–9
Majumdar A, Singhal V (2017) Noisy deep dictionary learning: application to Alzheimer’s Disease classification. In: Neural networks (IJCNN), 2017 international joint conference on. IEEE, pp 2679–2683
Avendi MR, Kheradvar A, Jafarkhani H (2016) A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 30:108–119
Havaei M, Guizard N, Larochelle H, Jodoin PM (2016) Deep learning trends for focal brain pathology segmentation in MRI. In: Machine learning for health informatics. Springer, Cham, pp 125–148
Andreu-Perez J, Poon CCY, Merrifield RD, Wong STC, Yang GZ (2015) Big data for health. IEEE J Biomed Heal Inform 19(4):1193–1208
Rahhal MMA, Bazi Y, Alhichri H, Alajlan N, Melgani F, Yager RR (2016) Deep learning approach for active classification of electrocardiogram signals. Inf Sci (Ny) 345:340–354
Liang Z, Zhang G, Huang JX, Hu QV (2014) Deep learning for healthcare decision making with EMRs. In: Bioinformatics and Biomedicine (BIBM), 2014 IEEE International conference on. IEEE, pp 556–559
Futoma J, Morris J, Lucas J (2015) A comparison of models for predicting early hospital readmissions. J Biomed Inform 56:229–238
Yan Y, Qin X, Wu Y, Zhang N, Fan J, Wang L (2015) A restricted Boltzmann machine based two-lead electrocardiography classification. In: 2015 IEEE 12th international conference on wearable and implantable body sensor networks (BSN)
Che Z, Purushotham S, Khemani R, Liu Y (2015) Distilling knowledge from deep networks with applications to healthcare domain. arXiv preprint arXiv:1512.03542
Huang T, Lan L, Fang X, An P, Min J, Wang F (2015) Promises and challenges of big data computing in health sciences. Big Data Res 2(1):2–11
Ong BT, Sugiura K, Zettsu K (2016) Dynamically pre-trained deep recurrent neural networks using environmental monitoring data for predicting PM2.5. Neural Comput. Appl 27(6):1553–1566
Zhao L, Chen J, Chen F, Wang W, Lu CT, Ramakrishnan N (2016) SimNest: social media nested epidemic simulation via online semi-supervised deep learning. Proc IEEE Int Conf Data Min i:639–648
Han S, Mao H, Dally WJ (2015) Deep compression: compressing deep neural networks with pruning, trained quantization and Huffman coding. arXiv preprint arXiv:1510.00149
Kim Y-D et al (2015) Compression of deep convolutional neural networks for fast and low power mobile applications. arXiv preprint arXiv:1511.06530
Lin Y et al (2017) Deep gradient compression: Reducing the communication bandwidth for distributed training. arXiv preprint arXiv:1712.01887
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pandey, S.K., Janghel, R.R. Recent Deep Learning Techniques, Challenges and Its Applications for Medical Healthcare System: A Review. Neural Process Lett 50, 1907–1935 (2019). https://doi.org/10.1007/s11063-018-09976-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-018-09976-2