Abstract
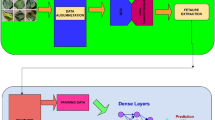
In this paper, a lightweight convolutional neural network is proposed for the classification of plant diseases, containing 63 classes of states for 11 plant species. The different context of experimental data and data in the real environment, insufficient accuracy of the model classification, and oversized model are three main problems of deep learning techniques applied to agricultural production. In this paper, we mainly focus on these three problems. First, the GrabCut algorithm is adopted to unify the background of the experimental data and the real data to black, allowing the trained model to have the same good effect when applied in practice. Then, we propose a new coordinate attention block to improve the classification accuracy of convolutional neural networks and empirically demonstrate the effectiveness of our approach with several state-of-the-art CNN models. Finally, channel pruning is applied to the trained model, which reduces the model size and computational effort by 85.19\(\%\) and 92.15\(\%\) respectively with little change in the model accuracy, making it better suited for agricultural platforms with lower memory and computational capacity.








Similar content being viewed by others
References
Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A (2019) The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3(3):430–439
Shrivastava VK, Pradhan MK (2020) Rice plant disease classification using color features: a machine learning paradigm. J Plant Pathol 103:17
Xie C, Yong H (2016) Spectrum and image texture features analysis for early blight disease detection on eggplant leaves. Sensors 16(5):676
Yang X, Zhang R, Zhai Z, Pang Y, Jin Z (2019) Machine learning for cultivar classification of apricots (Prunus armeniaca l.) based on shape features: sciencedirect. Sci Hortic 256:108524–108524
Fang W, Ding YA, Zhang FA, Sheng V (2019) Dog: a new background removal for object recognition from images. Neurocomputing 361:85–91
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419. https://doi.org/10.3389/fpls.2016.01419
Hang J, Zhang D, Chen P, Zhang J, Wang B (2019) Classification of plant leaf diseases based on improved convolutional neural network. Sensors. https://doi.org/10.3390/s19194161
Yan Q, Yang B, Wang W, Wang B, Chen P, Zhang J (2020) Apple leaf diseases recognition based on an improved convolutional neural network. Sensors. https://doi.org/10.3390/s20123535
Khatkar BS, Chaudhary N, Dangi P (2016) Production and consumption of grains: India
Rother C (2004) Grabcut : interactive foreground extraction using iterated graph cuts. Proc Siggraph 23:3
Hou Q, Zhou D, Feng J (2021) Coordinate attention for efficient mobile network design. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp 13713–13722
Zhuang L, Li J, Shen Z, Gao H, Zhang C (2017) Learning efficient convolutional networks through network slimming. In: 2017 IEEE international conference on computer vision (ICCV)
Sardogan M, Tuncer A, Ozen Y (2018) Plant leaf disease detection and classification based on cnn with lvq algorithm. In: 2018 3rd international conference on computer science and engineering (UBMK)
Mahmoud F, Haines D, Al-Ozairi E, Dashti A (2016) Effect of black tea consumption on intracellular cytokines, regulatory t cells and metabolic biomarkers in type 2 diabetes patients. Phytother Res 30(3):454–462
Saric S, Notay M, Sivamani RK (2017) Green tea and other tea polyphenols: effects on sebum production and acne vulgaris. Antioxidants 6(1):2
Tangney CC, Rasmussen HE (2013) Polyphenols, inflammation, and cardiovascular disease. Curr Atheroscler Rep 15(5):324
Alshatwi AA, Al Obaaid MA, Al Sedairy SA, Ramesh E, Lei KY (2011) Black and green tea improves lipid profile and lipid peroxidation parameters in wistar rats fed a high-cholesterol diet. J Physiol Biochem 67(1):95–104
Shang X, Song M, Yu C (2019) Hyperspectral image classification with background. In: IGARSS 2019-2019 IEEE international geoscience and remote sensing symposium, 2714–2717 .IEEE
Boykov Y, Kolmogorov V (2003) Computing geodesics and minimal surface via graph cuts. IEEE
Chen L.C, Papandreou G, Schroff F, Adam H (2017) Rethinking atrous convolution for semantic image segmentation
Douillard A, Chen Y, Dapogny A, Cord M (2020) Plop: learning without forgetting for continual semantic segmentation
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Zagoruyko S, Komodakis N (2016) Wide residual networks. arXiv preprint arXiv:1605.07146
Xie S, Girshick R, Dollár P, Tu Z, He K.(2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 1492–1500
Cao Y, Xu J, Lin S, Wei F, Hu H (2019) Gcnet: Non-local networks meet squeeze-excitation networks and beyond. In: Proceedings of the IEEE/CVF international conference on computer vision workshops, 0–0
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H (2019) Dual attention network for scene segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 3146–3154
Hu H, Gu J, Zhang Z, Dai J, Wei Y (2018) Relation networks for object detection. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, 3588–3597
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, 7132–7141
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV). pp 3–19
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 770–778
Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International conference on machine learning, 6105–6114. PMLR
Ma N, Zhang X, Zheng H-T, Sun J (2018) Shufflenet v2: practical guidelines for efficient cnn architecture design. In: Proceedings of the European conference on computer vision (ECCV). pp 116–131
Howard A, Sandler M, Chu G, Chen L.-C, Chen B, Tan M, Wang W, Zhu Y, Pang R, Vasudevan V, et al (2019) Searching for mobilenetv3. In: Proceedings of the IEEE/CVF international conference on computer vision. pp 1314–1324
Han K, Wang Y, Tian Q, Guo J, Xu C, Xu C (2020) Ghostnet: more features from cheap operations. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp 1580–1589
Han S, Pool J, Tran J, Dally WJ (2015) Learning both weights and connections for efficient neural networks
Wen W, Wu C, Wang Y, Chen Y, Li H (20196) Learning structured sparsity in deep neural networks
Kumar A, Shaikh AM, Li Y, Bilal H, Yin B (2021) Pruning filters with l1-norm and capped l1-norm for cnn compression. Appl Intell 51:1152–1160
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Bach F, Blei, D. (eds.), Proceedings of the 32nd international conference on machine learning. Proceedings of machine learning research, 37: 448–456. PMLR, Lille, France. http://proceedings.mlr.press/v37/ioffe15.html
Acknowledgements
This work is supported by China Agriculture Research System of MOF and MARA, the Project of Scientific and Technological Innovation Planning of Hunan Province (2020NK2008), Hunan Province Modern Agriculture Technology System for Tea Industry, the National Natural Science Foundation of China (42130716). We are grateful to the High Performance Computing Center of Central South University for partial support of this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, F., Wang, Y. & Tang, Z. Lightweight Plant Disease Classification Combining GrabCut Algorithm, New Coordinate Attention, and Channel Pruning. Neural Process Lett 54, 5317–5331 (2022). https://doi.org/10.1007/s11063-022-10863-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-022-10863-0