Abstract
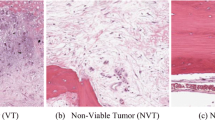
Out of the various types of primary bone cancers, Osteosarcoma is one of the most common malignant bone tumors. Children and teenagers are mostly affected by Osteosarcoma, which weakens the strength of their bones and sometimes may even result in death. It is important to develop an intelligent classifier that detect osteosarcoma accurately so that proper treatment can be given to the patient timely. In this paper, we propose an intelligent classifier that classifies osteosarcoma whole slide images (WSIs) into Viable Tumor, Non-Viable Tumor and Non-Tumor. To extract the region of interest (ROI) from WSIs, a Multi-Feature Non-Seed-based Region Growing algorithm (MFNSRG) based on intra-region homogeneity and inter-region heterogeneity maximization is used. We use textural heterogeneity along with color heterogeneity as the matching criteria during region growing. Finally, the background is eliminated using thresholding based on the size of a region and the ROI is obtained. The performance of MFNSRG is further improved by using Marine Predators, a recently proposed metaheuristic algorithm, which is used to obtain optimal value of segmentation parameters. Here, we use handcrafted methods to extract relevant features from the segmented image which are given as input to the classifier. The results of experimentation prove the superiority of the proposed approach as compared to the existing state-of-the-art algorithms.











Similar content being viewed by others
References
Lindsey BA, Markel JE, Kleinerman ES (2017) Osteosarcoma overview. Rheumatol Therapy 4(1):25–43
Durfee RA, Mohammed M, Luu HH (2016) Review of osteosarcoma and current manaement. Rheumatol Therapy 3(2):221–243
Ottaviani G, Jaffe N (2010) The Epidemiology of Osteosarcoma. In: Jaffe N, Bruland O, Bielack S (eds) Pediatric and Adolescent Osteosarcoma. Cancer Treatment and Research, Springer, Boston, MA
Martin E, Senders JT, ter Wengel PV (2019) Treatment and survival of osteosarcoma and Ewing sarcoma of the skull: a SEER database analysis. Acta Neurochir 161:317–325
Mirabello L, Troisi RJ, Savage SA (2009) Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 115(7):1531–1543. https://doi.org/10.1002/cncr.24121
Goode A, Gilbert B, Harkes J, Jukic D, Satyanarayanan M (2013) OpenSlide: A vendor-neutral software foundation for digital pathology. J Pathol Informat 4:1–27
Misaghi A, Goldin A, Awad M, Kulidjian AA (2018) Osteosarcoma: a comprehensive review. SICOT-J 4:12
Mishra R, Daescu O, Leavey P, Rakheja D, Sengupta A (2018) Convolutional neural network for histopathological analysis of osteosarcoma. J Comput Biol 25(3):313–325
Arunachalam HB, Mishra R, Armaselu B, Daescu O, Martinez M, Leavey PJ, Rakheja D, Cederberg KB, Sengupta AL, Ni’suilleabhain M (2017) Computer aided image segmentation and classification for viable and non-viable tumor identification in osteosarcoma. Pacific Symposium on Biocomputing 195–206
Leavey P, Sengupta A, Rakheja D, Daescu O, Arunachalam HB, Mishra R (2019) Osteosarcoma data from UT Southwestern/UT Dallas for Viable and Necrotic Tumor Assessment . The Cancer Imaging Archive
Wang H, Oliensis J (2010) Generalizing edge detection to contour detection for image segmentation. Comput Vis Image Underst 114:731–744
Gruenwedel S, Van Hese P, Philips W (2011) An Edge-Based Approach for Robust Foreground Detection. In: Blanc-Talon J., Kleihorst R., Philips W., Popescu D., Scheunders P. eds Advanced Concepts for Intelligent Vision Systems ACIVS 2011 Lecture Notes in Computer Science, Springer, Berlin, Heidelberg
Adams R, Bischof L (1994) Seeded region growing. IEEE Trans Pattern Anal Mach Intell 16(6):641–647
Wu L, Wang Y, Long J, Liu Z (2015) A Non-seed-based Region Growing Algorithm for High Resolution Remote Sensing Image Segmentation. In: Zhang YJ (ed) Image and Graphics ICIG 2015 Lecture Notes in Computer Science. Springer, Cham
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Haniza Y, Hamzah A (2013) Gradient based adaptive thresholding. J Vis Commun Image Represent 24(7):926–936
Qureshi MN, Ahamad MV (2018) An improved method for image segmentation using K-means clustering with neutrosophic logic. Procedia Comput Sci 132:534–540
Priyadarsan P, Bhoi N (2018) Fuzzy clustering based transition region extraction for image segmentation. Eng Sci Technol Int J 21(4):547–563
Faramarzi A, Heidarinejad M, Mirjalili S, Gandomi A (2020) Marine Predators Algorithm: A Nature-inspired Metaheuristic. Expert Syst Applications 152:11337
Yusufiyah HKN, Nugroho HA, Adji TB, Nugroho (2015) A Feature extraction for classifying lesion’s shape of breast ultrasound images. 2nd International Conference on Information Technology, Computer, and Electrical Engineering (ICITACEE), Semarang, Indonesia, 102–106
Rahmawaty M, Nugroho HA, Triyani Y, Ardiyanto I, Soesanti I (2016) Classification of breast ultrasound images based on texture analysis. International Conference on Biomedical Engineering (iBioMed) 1–6
Bakheet S (2017) An SVM framework for malignant melanoma detection based on optimized HOG features. Computation 5(1):4
Solmaz A, Tajeripour F (2016) Detection of Brain Tumor in 3D MRI Images using Local Binary Patterns and Histogram Orientation Gradient. Neurocomputing 219
Razzaq S, Mubeen N, Kiran U, Asghar MA, Fawad F (2020) Brain Tumor Detection from MRI Images Using Bag of Features and Deep Neural Network. International Symposium on Recent Advances in Electrical Engineering & Computer Sciences (RAEE & CS) 1–6
Liu D, Gan T, Rao N, Xu G, Zeng B, Li H (2015) Automatic detection of early gastrointestinal cancer lesions based on optimal feature extraction from gastroscopic images. J Med Imag Health Inf 5(2):296
Li Y, Deng L, Yang X (2019) Early diagnosis of gastric cancer based on deep learning combined with the spectral-spatial classification method. Biomed Opt Express 10(10):4999–5014
Bisla D, Choromanska A, Berman RS, Stein JA, Polsky D (2019) Towards automated melanoma detection with deep learning: Data purification and augmentation. In Proceedings - 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, CVPRW 2019, 2720–2728
Cao Z, Duan L, Yang G (2019) An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures. BMC Med Imaging 19:51
Chang J, Yu J, Han T, Chang HJ, Park E (2017) A method for classifying medical images using transfer learning: A pilot study on histopathology of breast cancer. In 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services, Healthcom 2017, 1–4
Tosta TAA, Neves LA, do Nascimento MZ, (2017) Segmentation methods of H&E-stained histological images of lymphoma: a review. Informatics in Medicine Unlocked 9:35–43
Lan C, Heindl A, Huang X (2015) Quantitative histology analysis of the ovarian tumour microenvironment. Sci Rep 5:16317
Mishra R, Daescu O, Leavey P, Rakheja D, Sengupta A (2017) Histopathological Diagnosis for Viable and Non-viable Tumor Prediction for Osteosarcoma Using Convolutional Neural Network International Symposium on Bioinformatics Research and Applications. Springer, Cham
Arunachalam HB, Mishra R, Daescu O (2019) Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PLoS ONE 14(4):23
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet Classification with Deep Convolutional Neural Networks. Neural Information Processing Systems
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation applied to handwritten zip code recognition. Neural Comput 1(4):541–551
Anisuzzaman DM, Barzekar H, Tong L, Luo J, Yu Z (2020) A Deep Learning Study on Osteosarcoma Detection from Histological Images. https://arxiv.org/pdf/2011.01177.pdf
Fan J, Yau DKY, Elmagarmid A, Aref W (2001) Image segmentation by integrating color edge detection and seeded region growing. IEEE Trans Image Process 10(10):1454–1466
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24(7):971–987
Fotheringham AS, Brunsdon C, Charlton M (2000) Quantitative Geography: Perspectives on Spatial Analysis. Sage, London
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bansal, P., Singhal, A. & Gehlot, K. Osteosarcoma Detection from Whole Slide Images Using Multi-Feature Non-Seed-Based Region Growing Segmentation and Feature Extraction. Neural Process Lett 55, 3671–3693 (2023). https://doi.org/10.1007/s11063-022-10914-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-022-10914-6