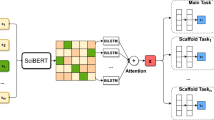
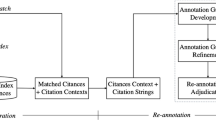
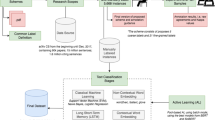
Abstract
A study on scientific data citation is crucial to promote data sharing and is the basis for the examination of scientific data measurement and analysis. To this end, it is necessary to identify and label data reference information. Currently, there are many supervised methods for entity recognition and relationship extraction of diseases, drugs, proteins, symptoms, etc., but they have not discussed the effectiveness of scientific data recognition. To fill this gap, the effectiveness of the classical machine learning model and the deep learning model on recognizing scientific data citation are discussed in this study. In experiments, this study took the full text of scientific and technical papers as the research object, conducted annotated citation classification based on rules and manual recognition of their references to form a dataset. The results of the empirical study showed that: (1) the methods used in this paper can achieve automatic identification and extraction of data citations and can address the problem of automating the construction of citation relationships between scientific and technical literature and scientific data; (2) the BERT-based models have the optimal effectiveness in the recognition task of scientific data citation, especially the BioBERT and SciBERT; (3) the full-text information has a crucial impact on the recognition results.






Similar content being viewed by others
References
Beltagy, I., Lo, K., & Cohan, A. (2019). SciBERT: A pretrained language model for scientific text. arXiv preprint arXiv:1903.10676.
Bengio, Y., Ducharme, R., Vincent, P., & Jauvin, C. (2003). A neural probabilistic language model. Journal of Machine Learning Research, 3(6), 1137–1155. https://doi.org/10.1162/153244303322533223
Borgman, C. L. (2015). Scholarship in the Networked World: Big Data, Little Data, noData[R]. University of California. https://escholarship.org/uc/item/38v6n99v
Carletta, J. (1996). Assessing agreement on classification tasks: The kappa statistic. Computational Linguistics, 22(2), 249–254.
Chapman, B., & Chang, J. (2000). Biopython: Python tools for computational biology. ACM SIGBIO Newsletter, 20, 15–19. https://doi.org/10.1145/360262.360268
Cui, B.-G., & Chen, X. (2010). An improved hidden markov model for literature metadata extraction (Vol. 6215). Springer.
Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2019). BERT: Pre-training of deep bidirectional transformers for language understanding, Minneapolis, Minnesota.
Duke, M., & Ball, A. (2012, 2012/10/30). How to Cite Datasets and Link to Publications. Paper presented at the 23rd International CODATA Conference.
Ghavimi, B., Mayr, P., Vahdati, S., & Lange, C. (2016). Identifying and improving dataset references in social sciences full texts. arXiv preprint arXiv:1603.01774.
Grechkin, M., Poon, H., & Howe, B. (2017). Wide-Open: Accelerating public data release by automating detection of overdue datasets. Plos Biology, 15(6), e2002477.
Green, T. (2009). OECD publishing white paper we need publishing standards for datasets and data tables. Learned Publishing. https://doi.org/10.1087/20090411
Haeussler, M., Gerner, M., & Bergman, C. M. (2011). Annotating genes and genomes with DNA sequences extracted from biomedical articles. Bioinformatics, 27(7), 980–986. https://doi.org/10.1093/bioinformatics/btr043
Henderson, T., & Kotz, D. (2015). Data citation practices in the CRAWDAD wireless network data archive. D-Lib Magazine. https://doi.org/10.1045/january2015-henderson
Hou, L., Zhang, J., Wu, O., Yu, T., Wang, Z., Li, Z., Gao, J., Ye, Y., & Yao, R. (2020). Method and dataset entity mining in scientific literature: A CNN + Bi-LSTM model with self-attention. Knowledge-Based Systems. https://doi.org/10.1016/j.knosys.2021.107621
Kim, Y. (2014). Convolutional neural networks for sentence classification. http://arxiv.org/abs/1408.5882. Retrieved from https://ui.adsabs.harvard.edu/abs/2014arXiv1408.5882K
Lai, S. W., Xu, L. H., Liu, K., & Zhao, J. (2015). Recurrent convolutional neural networks for text classification. Proceedings of the Twenty-Ninth Aaai Conference on Artificial Intelligence, pp. 2267–2273.
Lan, Z., Chen, M., Goodman, S., Gimpel, K., Sharma, P., & Soricut, R. (2019). ALBERT: A Lite BERT for Self-supervised Learning of Language Representations.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234–1240. https://doi.org/10.1093/bioinformatics/btz682
Major, G. R. (2011). Impact of NASA EOS instrument data on the scientific literature: 10 years of published research results from Terra, Aqua, and Aura. Issues in Science and Technology Librarianship. https://doi.org/10.5062/F4CC0XMJ
Mikolov, T., Sutskever, I., Chen, K., Corrado, G., & Dean, J. (2013). Distributed representations of words and phrases and their compositionality. http://arxiv.org/abs/1310.4546. Retrieved from https://ui.adsabs.harvard.edu/abs/2013arXiv1310.4546M
Mooney, H. (2011). Citing data sources in the social sciences: Do authors do it? Learned Publishing, 24, 99–108. https://doi.org/10.1087/20110204
Mooney, H., & Newton, M. P. (2012). The anatomy of a data citation: Discovery, reuse, and credit. Journal of Librarianship and Scholarly Communication. https://doi.org/10.7710/2162-3309.1035
Neveol, A., Wilbur, W. J., & Lu, Z. Y. (2011). Extraction of data deposition statements from the literature: A method for automatically tracking research results. Bioinformatics, 27(23), 3306–3312. https://doi.org/10.1093/bioinformatics/btr573
Park, H., You, S., & Wolfram, D. (2017). Is informal data citation for data sharing and re-use more common than formal data citation? Proceedings of the Association for Information Science and Technology, 54, 768–769. https://doi.org/10.1002/pra2.2017.14505401150
Park, H., You, S., & Wolfram, D. (2018). Informal data citation for data sharing and reuse is more common than formal data citation in biomedical fields. Journal of the Association for Information Science and Technology. https://doi.org/10.1002/asi.24049
Peters, I., Kraker, P., Lex, E., Gumpenberger, C., & Gorraiz, J. (2015). Research data explored: Citations versus Altmetrics. http://arxiv.org/abs/1501.03342. Retrieved from https://ui.adsabs.harvard.edu/abs/2015arXiv150103342P
ESIP Data Preservation and Stewardship Committee. 2019. Data Citation Guidelines for Earth Science Data. Ver. 2. Earth Science Information Partners.https://doi.org/10.6084/m9.figshare.8441816
Sokolova, M., & Lapalme, G. (2009). A systematic analysis of performance measures for classification tasks. Information Processing & Management, 45(4), 427–437. https://doi.org/10.1016/j.ipm.2009.03.002
Tenopir, C., Allard, S., Douglass, K., Aydinoglu, A. U., Wu, L., Read, E., Manoff, M., & Frame, M. (2011). Data sharing by scientists: practices and perceptions. PLoS ONE, 6, e21101. https://doi.org/10.1371/journal.pone.0021101
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J. W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., & Mons, B. (2016). Comment: The FAIR guiding principles for scientific data management and stewardship. Scientific Data. https://doi.org/10.1038/sdata.2016.18
Yu, Q., Ding, Y., Song, M., Song, S. J., Liu, J. H., & Zhang, B. (2015). Tracing database usage: Detecting main paths in database link networks. Journal of Informetrics, 9(1), 1–15. https://doi.org/10.1016/j.joi.2014.10.002
Zenk-Moltgen, W., & Lepthien, G. (2014). Data sharing in sociology journals. Online Information Review, 38(6), 709–722. https://doi.org/10.1108/Oir-05-2014-0119
Zhang, Q., Cheng, Q., & Lu, W. (2016). A bootstrapping-based method to automatically identify data-usage statements in publications. Journal of Data and Information Science, 1, 1–17. https://doi.org/10.20309/jdis.201606
Zhao, M. N., Yan, E. J., & Li, K. (2018). Data set mentions and citations: A content analysis of full-text publications. Journal of the Association for Information Science and Technology, 69(1), 32–46. https://doi.org/10.1002/asi.23919
Acknowledgements
We would like to thank the anonymous reviewers for their helpful suggestions.
Author information
Authors and Affiliations
Contributions
NY: Concieved and designed the analysis; Collected the data; Contributed data or analysis tool; Performed the analysis; Wrote the paper. ZZ: Concieved and designed the analysis; Wrote the paper. FH: Performed the analysis; Wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, N., Zhang, Z. & Huang, F. A study of BERT-based methods for formal citation identification of scientific data. Scientometrics 128, 5865–5881 (2023). https://doi.org/10.1007/s11192-023-04833-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-023-04833-z