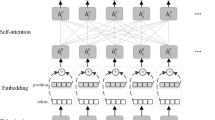
Abstract
The accumulation and explosive growth of the electronic medical records (EMRs) make the name entity recognition (NER) technologies become critical for the meaningful use of EMR data and then the practice of evidence-based medicine. The dominate NER approaches use the distributed representation of the words and characters to build deep learning-based NER models. However, for the task of biomedical named entity recognition, there are a large amount of complicated medical terminologies that are composed of multiple words. Splitting these terminologies to learn the word and character embeddings might cause semantic ambiguities. In this paper, we treat each medical terminology as a concept and propose a concept-enhanced named entity recognition model (CNER), where the features from three different granularities (i.e., concept, word, and character) are combined together for bio-NER. The extensive experiments are conducted on two real-world corpora: fully labeled corpus and partially labeled corpus. CNER achieves the highest F1 score (fully labeled corpus: precision = 88.23, recall = 88.29, and F1 = 88.26; partially labeled corpus: precision = 87.03, recall = 88.19, and F1 = 87.61) by outperforming the baseline CW-BLSTM-CRF approach for 0.58% and 1.15% respectively, which demonstrates the effectiveness of the proposed approach.





Similar content being viewed by others
References
Li J, Liu C, Liu B, Mao R, Chen S, Pan H, Wang Q (2015) Diversity-aware retrieval of medical records. Comput Ind 69(1):30–39
Yang L, Zhou Y (2014) Exploring feature sets for two-phase biomedical named entity recognition using semi-CRFs. Knowl Inf Syst 40(2):439–453
Chowdhury S, Dong X, Qian L (2018) A multitask bi-directional RNN model for named entity recognition on Chinese electronic medical records. BMC Bioinform 19(17):499–513
Yonghui W, Jiang M, Lei J (2015) Named entity recognition in Chinese clinical text using deep neural network. Stud Health Technol Inf 22(8):321–342
Li J, Zhao Y, Liu B (2012) Exploiting semantic resources for large scale text categorization. J Intell Inf Syst 39(3):763–788
Seifollahi S, Shajari M (2019) Word sense disambiguation application in sentiment analysis of news headlines: an applied approach to FOREX market prediction. J Intell Inf Syst 52(1):57–83
Nguyen DQ, Verspoor K (2019) From POS tagging to dependency parsing for biomedical event extraction. BMC Bioinform 20(2):78–101
Pandey AC, Rajpoot DS, Saraswat M (2017) Twitter sentiment analysis using hybrid cuckoo search method. Inf Process Manag 12(3):565–576
Yang Z, Chen W, Wang F, Xu B (2019) Effectively training neural machine translation models with monolingual data. Neurocomputing 333:240–247
Wu H, Li J, Kang Y, Zhong T (2018) Exploring noise control strategies for UMLS-based query expansion in health and biomedical information retrieval. J Ambient Intell Humanized Comput. https://doi.org/10.1007/s12652-018-0836-x
Zhao Q, Kang YY, Li JQ, Wang D (2018) Exploiting the semantic graph for the representation and retrieval of medical documents. Comput Biol Med 101:39–50
Lample G, Ballesteros M et al (2016) Neural architectures for named entity recognition. Association for Computational Linguistics, Stroudsburg, pp 260–270
Marrero M, Urbano J, Sánchez-Cuadrado S, Morato J, Gómez-Berbís M (2013) Named entity recognition: fallacies, challenges and opportunities. Comput Stand Interfaces 35(5):482–489
Zhang S, Elhadad N (2013) Unsupervised biomedical named entity recognition: experiments with clinical and biological texts. J Biomed Inform 46(6):1088–1098
Chen X, Xu L, Liu Z, Sun M, Luan H (2015) Joint learning of character and word embeddings. In: International Joint Conference on Artificial Intelligence (IJCAI), pp 1236–1242
Chiu JPC, Nichols E (2016) Named entity recognition with bidirectional LSTM-CNNs. Comput Sci 24(8):261–273
Anthony P, Alfred R, Leong LC (2013) A rule-based named-entity recognition for malay articles. In: International Conference on Advanced Data
Zaghouani W (2012) RENAR: a rule-based arabic named entity recognition system. In: ACM Transactions on Asian Language Information
Leong ASC, Dey S, Evans JS (2007) Probability of error analysis for hidden Markov model filtering with random packet loss. IEEE Trans Signal Process 55(3):218–231
Xiao J, Wang X, Liu B (2001) The study of a nonstationary maximum entropy Markov model and its application on the pos-tagging task. ACM Trans Asian Lang Inf 32(5):334–345
Furey TS, Cristianini N, Duffy N (2000) Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 12(9):189–202
Zheng S, Jayasumana S, Romera-Paredes B, Vineet V et al (2015) Conditional random fields as recurrent neural networks. In: IEEE International Conference on Computer Vision (ICCV), vol 32(6), pp 206–015
Li C, Song R, Liakata M, Vlachos A, Seneff S, Zhang X (2015) Using word embedding for bioevent extraction. In: Proceedings of the 2015 Workshop on Biomedical Natural Language Processing (BioNLP 2015). Association for Computational Linguistics, Stroudsburg, pp 121–126
Lei J, Tang B, Lu X, Gao K, JiangM X (2013) A comprehensive study of named entity recognition in Chinese clinical text. J Am Med Inform Assoc 21(5):808–814
Gu J, Wang Z, Kuen J et al (2015) Recent advances in convolutional neural networks. Comput Sci 41(12):321–324
Shen Y, Wang J (2008) An improved algebraic criterion for global exponential stability of recurrent neural networks with time-varying delays. IEEE Trans Neural Netw 37(9):523–535
Yao C, Qu Y, Jin B, Guo L, Li C, Cui W, Feng L (2016) A convolutional neural network model for online medical guidance. IEEE Access 4:4094–4103
Sahu SK, Anand A (2016) Recurrent neural network models for disease name recognition using domain invariant features. arXiv preprint arXiv:1606.09371
Wu Y, JiangM LJ, Xu H (2015) Named entity recognition in chinese clinical text using deep neural network. Stud Health Technol Inform 216:624
Chiu JP, Nichols E (2015) Named entity recognition with bidirectional LSTM-CNNS. arXiv preprint arXiv:1511.08308
Lei J, Tang B, Lu X, Gao K, JiangM X (2013) A comprehensive study of named entity recognition in chinese clinical text. J Am Med Inform Assoc 21(5):808–814
Wang X, Li J, Tan Z, Ma L, Li F, Huang M (2016) The state of the art and future tendency of “Internet+” oriented network technology. J Comput Res Dev 53(4):729–741
Dong X, Qian L, Guan Y, Huang L, Yu Q, Yang J (2016) A multiclass classification method based on deep learning for named entity recognition in electronic medical records. In: Scientific Data Summit (NYSDS), IEEE, New York, pp 1–10
Wang P, Qian Y, Soong FK He L, Zhao H (2015) A unified tagging solution: bidirectional LSTM recurrent neural network with word embedding. arXiv preprint arXiv:1511.00215
Zhao Z, Yang Z, Luo L, Zhang Y, Wang L, Lin H, Wang J (2016) ML-CNN: a novel deep learning based disease named entity recognition architecture. In: 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp 794–794
Hochreiter S, Schmidhuber J (1997) Long short-term memory. Neural Comput 9(8):1735–1780
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(1):192–200
Forney GD Jr (1973) The Viterbi algorithm. In: Proceedings of the IEEE, vol 4, pp 413–426
Han X, Ruonan R (2011) The method of medical named entity recognition based on semantic model and improved SVM–KNN algorithm. In: Seventh International Conference on Semantics Knowledge and Grid, vol 12, pp 146–155
Li J, Liu C (2012) A cooperative co-learning approach for concept detection in documents. In: IEEE Sixth International Conference on Semantic Computing, 25 Oct 2012
Fridsma D (2012) Electronic health records: the HHS perspective. Computer 45(11):24–26. https://doi.org/10.1109/MC.2012.371
Acknowledgements
This study is supported by the Xinjiang Nature Science Foundation of China (2019D01A23).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, Q., Wang, D., Li, J. et al. Exploiting the concept level feature for enhanced name entity recognition in Chinese EMRs. J Supercomput 76, 6399–6420 (2020). https://doi.org/10.1007/s11227-019-02917-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-019-02917-3