Abstract
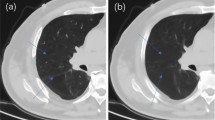
This work aimed to explore chest computed tomography (CT) image segmentation of patients with large-cell neuroendocrine carcinoma (LCNEC) based on deep learning, as well as the clinical manifestations and imaging and pathological features of LCNEC patients. Clinical data of 40 patients with LCNEC confirmed by pathological examination in the X Hospital from December 2015 to December 2017 were retrospectively selected. CT image data were segmented by TJ-1 model modified full convolutional neural network (FCNN) model. The accuracy and training time of TJ-1 FCNN model and classic deep learning segmentation network model AlexNet model were compared in terms of image segmentation. According to the image segmentation results by TJ-1 FCNN model, chest CT images of LCNEC patients, were reviewed, and the clinical manifestations, as well as the imaging and pathological features of the patients were reviewed, sorted, and summarized. The results showed that the image segmentation accuracy of TJ-1 network model (99.38%) was higher than that of AlexNet model. The iteration training time of TJ-1 network model for 30 times was 45 min, lower than that of AlexNet model (82 min). LCNEC was more likely to be found in elderly male with a long history of smoking. The clinical symptoms were cough, sputum, sputum blood, and chest pain with no significant specificity. CT imaging showed that peripheral mass was the most common manifestation (67.5%), both lungs were visible, the upper lobe was more likely with lesion (60%), the edge of the lesion was clear or smooth (57.5%), which was lobulated (70%). Under the light microscope, tumor cells were characterized by large volume, low nucleocytoplasmic ratio, high mitosis, and large area necrosis. The positive rates of immunohistochemical neuroendocrine markers were CD56 (62.5%), CgA (50%), and Syn (85%), among which Syn was with the highest positive rate. To sum up, LCNEC lacked clinical and radiological specificity manifestations, while chest CT image segmentation based on TJ-1 FCNN model can quickly mark the location of the lesion, providing technical support for the diagnosis and evaluation of LCNEC clinically.











Similar content being viewed by others
References
Iyoda A, Makino T, Koezuka S et al (2014) Treatment options for patients with large cell neuroendocrine carcinoma of the lung. Gen Thorac Cardiovasc Surg 62:351–356. https://doi.org/10.1007/s11748-014-0379-9
Lo Russo G, Pusceddu S, Proto C (2016) Treatment of lung large cell neuroendocrine carcinoma. Tumour Biol 37:7047–7057. https://doi.org/10.1007/s13277-016-5003-4
Sholl LM (2016) The molecular pathology of lung Cancer. Surg Pathol Clin 9:353–378. https://doi.org/10.1016/j.path.2016.04.003
Schnabel PA, Junker K (2015) Pulmonary neuroendocrine tumors in the new WHO 2015 classification: start of breaking new grounds? Pathologe 36:283–292. https://doi.org/10.1007/s00292-015-0030-2
He Y, Liu H, Wang S et al (2019) Prognostic nomogram predicts overall survival in pulmonary large cell neuroendocrine carcinoma. PLoS One 14:e0223275. https://doi.org/10.1371/journal.pone.0223275
Naidoo J, Santos-Zabala ML, Iyriboz T et al (2016) Large cell neuroendocrine carcinoma of the lung: clinico-pathologic features, treatment, and outcomes. Clin Lung Cancer 17:e121–e129. https://doi.org/10.1016/j.cllc.2016.01.003
Gálffy G (2018) Diagnosis and treatment of the neuroendocrine tumors of the lung. Magy Onkol 62:113–118
Zhang L, Wang X, Yang D et al (2020) Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans Med Imaging 39:2531–2540. https://doi.org/10.1109/TMI.2020.2973595
Christopher M, Bowd C, Belghith A, Goldbaum MH, Weinreb RN, Fazio MA, Girkin CA, Liebmann JM, Zangwill LM (2020) Deep learning approaches predict glaucomatous visual field damage from OCT optic nerve head en face images and retinal nerve fiber layer thickness maps. Ophthalmology 127(3):346–356. https://doi.org/10.1016/j.ophtha.2019.09.036
Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, Thng F, Peng L, Stumpe MC (2018) Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Pathol 42(12):1636–1646. https://doi.org/10.1097/PAS.0000000000001151
Zhang J, Xie Y, Wu Q, Xia Y (2019) Medical image classification using synergic deep learning. Med Image Anal 54:10–19. https://doi.org/10.1016/j.media.2019.02.010
Hesamian MH, Jia W, He X et al (2019) Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging 32:582–596. https://doi.org/10.1007/s10278-019-00227-x
Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O (2018) Deep learning with convolutional neural network in radiology. Jpn J Radiol 36(4):257–272. https://doi.org/10.1007/s11604-018-0726-3
Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P (2018) Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 1(95):43–54. https://doi.org/10.1016/j.compbiomed.2018.02.004
Noh KJ, Park SJ, Lee S (2019) Scale-space approximated convolutional neural networks for retinal vessel segmentation. Comput Methods Programs Biomed 178:237–246. https://doi.org/10.1016/j.cmpb.2019.06.030
Lu X, Chen Y, Li X (2018) Hierarchical recurrent neural hashing for image retrieval with hierarchical convolutional features. IEEE Trans Image Process 27(1):106–120. https://doi.org/10.1109/TIP.2017.2755766
Travis WD, Brambilla E, Nicholson AG et al (2015) The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10:1243–1260. https://doi.org/10.1097/JTO.0000000000000630
Weston AD, Korfiatis P, Kline TL et al (2019) Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 290:669–679. https://doi.org/10.1148/radiol.2018181432
Ma X, Hadjiiski LM, Wei J et al (2019) U-Net based deep learning bladder segmentation in CT urography. Med Phys 46:1752–1765. https://doi.org/10.1002/mp.13438
Bhardwaj A, Shah SBH, Shankar A et al (2020) Penetration testing framework for smart contract blockchain. Peer-to-Peer Netw Appl 4:1–16. https://doi.org/10.1007/s12083-020-00991-6
Kumar M, Alshehri M, AlGhamdi R et al (2020) A DE-ANN inspired skin cancer detection approach using fuzzy C-Means clustering. Mobile Netw Appl 25:1319–1329. https://doi.org/10.1007/s11036-020-01550-2
Qian Z, Hu Y, Zheng H et al (2016) Clinical analysis of 22 cases of pulmonary large cell neuroendocrine cancer. Zhongguo Fei Ai Za Zhi 19:82–87. https://doi.org/10.3779/j.issn.1009-3419.2016.02.05
Lee KW, Lee Y, Oh SW et al (2015) Large cell neuroendocrine carcinoma of the lung: CT and FDG PET findings. Eur J Radiol 84:2332–2338. https://doi.org/10.1016/j.ejrad.2015.07.033
Zombori T, Juhasz-Nagy G, Tiszlavicz L et al (2020) Large-cell neuroendocrine carcinoma of the lung - challenges of diagnosis and treatment. Orv Hetil 161:313–319. https://doi.org/10.1556/650.2020.31581
Hiroshima K, Mino-Kenudson M (2017) Update on large cell neuroendocrine carcinoma. Transl Lung Cancer Res 6:530–539. https://doi.org/10.21037/tlcr.2017.06.12
Deng C, Wu SG, Tian Y (2019) Lung large cell neuroendocrine carcinoma: an analysis of patients from the surveillance, epidemiology and end-results (SEER) database. Med Sci Monit 25:3636–3646. https://doi.org/10.12659/MSM.914541
Watanabe R, Ito I, Kenmotsu H et al (2013) Large cell neuroendocrine carcinoma of the lung: is it possible to diagnose from biopsy specimens? Jpn J Clin Oncol 43:294–304. https://doi.org/10.1093/jjco/hys221
Yang X, Lin D (2016) Changes of 2015 WHO histological classification of lung cancer and the clinical significance. Zhongguo Fei Ai Za Zhi 19:332–336. https://doi.org/10.3779/j.issn.1009-3419.2016.06.06
Chen M, Gong D (2019) Discrimination of breast tumors in ultrasonic images using an ensemble classifier based on TensorFlow framework with feature selection. J Investig Med 67(Suppl 1):A3. https://doi.org/10.1136/jim-2019-000994.9
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, C., Wang, X., Zhou, H. et al. Analysis of clinical features of large-cell neuroendocrine carcinoma patients guided by chest CT image under deep learning. J Supercomput 77, 9290–9307 (2021). https://doi.org/10.1007/s11227-021-03647-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-021-03647-1