Abstract
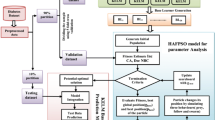
The progressive damage of kidney function and structure exhibits a high risk of cardiovascular disease and is the leading cause of life-threatening chronic kidney disease (CKD). Early diagnosing is the only way to prevent CKD from worsening. Different data mining and machine learning approaches are implemented for predicting chronic kidney disease; however, the extraction of hidden details from the clinical data is a difficult challenge. To overcome this situation, a new model for accurate prediction of chronic kidney disease is proposed in this article. This work evaluated the proposed predictive model using a CKD dataset. The raw chronic kidney disease dataset is initially preprocessed using data cleaning, data reduction as well as data transformation steps. These steps make the classification system easier to predict diseased and normal classes by enhancing the data quality. The features are then selected using a hybrid flash butterfly optimization algorithm. Here, the weight functions of the kernel soft plus extreme learning machine (KSELM) method are optimally tuned using the hybrid flash butterfly optimization (HFBO) algorithm to increase prediction accuracy. The efficiency of the proposed HFBO-KSELM algorithm is examined using different evaluation measures namely precision, accuracy, and specificity, recall, F1-score, and computation time. The results of the experimental analysis display superior performance of the proposed HFBO-KSELM algorithm over other existing methods, particularly with an accuracy of 97.8%, precision of 97.4%, recall of 96.9%, specificity of 97.5%, F1-score of 97.1%, computational time of 2.3 s, training accuracy of 0.978, testing accuracy of 0.94 and ROC/AUC of 0.97. Finally, the KSELM algorithm accurately predicts and classifies them into benign and malignant classes. The experimental result showed that the proposed HFBO-KSELM algorithm provides the effectiveness and robustness to detect chronic kidney disease.












Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Code availability
Not applicable.
References
Wang W, Chakraborty G, Chakraborty B (2020) Predicting the risk of chronic kidney disease (ckd) using machine learning algorithm. Appl Sci 11(1):202. https://doi.org/10.3390/app11010202
Salkar C (2021) A detailed analysis on kidney and heart disease prediction using machine learning. J Comput Nat Sci 1:9–14
Ghosh P, Shamrat FJM, Shultana S, Afrin S, Anjum AA, Khan AA (2020) Optimization of prediction method of chronic kidney disease using machine learning algorithm. In: 2020 15th International Joint Symposium on Artificial Intelligence and Natural Language Processing (iSAI-NLP), pp 1–6. https://doi.org/10.1109/iSAI-NLP51646.2020.9376787
Bhaskar N, Suchetha M (2021) A computationally efficient correlational neural network for automated prediction of chronic kidney disease. IRBM 42(4):268–276. https://doi.org/10.1016/j.irbm.2020.07.002
Al-Wahsh H, Lam NN, Quinn RR, Ronksley PE, Sood MM, Hemmelgarn B, Tangri N, Ferguson T, Tonelli M, Ravani P, Liu P (2022) Calculated versus measured albumin-creatinine ratio to predict kidney failure and death in people with chronic kidney disease. Kidney Int. https://doi.org/10.1016/j.kint.2022.02.034
Navaneeth B, Suchetha M (2020) A dynamic pooling based convolutional neural network approach to detect chronic kidney disease. Biomed Signal Process Control 62:102068. https://doi.org/10.1016/j.bspc.2020.102068
Berchtold L, Crowe LA, Combescure C, Kassaï M, Aslam I, Legouis D, Moll S, Martin PY, de Seigneux S, Vallée JP (2022) Diffusion-magnetic resonance imaging predicts decline of kidney function in chronic kidney disease and in patients with a kidney allograft. Kidney Int 101(4):804–813
Abdelaziz A, Salama AS, Riad AM, Mahmoud AN (2019) A machine learning model for predicting of chronic kidney disease based internet of things and cloud computing in smart cities. Security in smart cities: models, applications, and challenges. Springer, Cham, pp 93–114
Abdel-Fattah MA, Othman NA, Goher N (2022) Predicting chronic kidney disease using hybrid machine learning based on apache spark. Comput Intell Neurosci
Ren Y, Fei H, Liang X, Ji D, Cheng M (2019) A hybrid neural network model for predicting kidney disease in hypertension patients based on electronic health records. BMC Med Inform Decis Mak 19(2):131–138
Ma F, Sun T, Liu L, Jing H (2020) Detection and diagnosis of chronic kidney disease using deep learning-based heterogeneous modified artificial neural network. Futur Gener Comput Syst 111:17–26. https://doi.org/10.1016/j.future.2020.04.036
Khamparia A, Saini G, Pandey B, Tiwari S, Gupta D, Khanna A (2020) KDSAE: chronic kidney disease classification with multimedia data learning using deep stacked autoencoder network. Multim Tools Appl 79(47):35425–35440
Qin J, Chen L, Liu Y, Liu C, Feng C, Chen B (2019) A machine learning methodology for diagnosing chronic kidney disease. IEEE Access 8:20991–21002
Bhaskar N, Manikandan S (2019) A deep-learning-based system for automated sensing of chronic kidney disease. IEEE Sens Lett 3(10):1–4. https://doi.org/10.1109/LSENS.2019.2942145
Jerlin Rubini L, Perumal E (2020) Efficient classification of chronic kidney disease by using multi-kernel support vector machine and fruit fly optimization algorithm. Int J Imaging Syst Technol 30(3):660–673
Rubini LJ, Perumal E (2020) Hybrid kernel support vector machine classifier and grey wolf optimization algorithm based intelligent classification algorithm for chronic kidney disease. J Med Imaging Health Inf 10(10):2297–2307
Siddhartha M, Kumar V, Nath R (2022) Early-stage diagnosis of chronic kidney disease using majority vote–Grey Wolf optimization (MV-GWO). Heal Technol 12(1):117–136
Elhoseny M, Shankar K, Uthayakumar J (2019) Intelligent diagnostic prediction and classification system for chronic kidney disease. Sci Rep 9(1):1–14
Rady EHA, Anwar AS (2019) Prediction of kidney disease stages using data mining algorithms. Inf Med Unlocked 15:100178. https://doi.org/10.1016/j.imu.2019.100178
Pasadana IA, Hartama D, Zarlis M, Sianipar AS, Munandar A, Baeha S, Alam ARM (2019) Chronic kidney disease prediction by using different decision tree techniques. J Phys Conf Ser 1255(1):012024
Jongbo OA, Adetunmbi AO, Ogunrinde RB, Badeji-Ajisafe B (2020) Development of an ensemble approach to chronic kidney disease diagnosis. Sci Afr 8:e00456. https://doi.org/10.1016/j.sciaf.2020.e00456
Chaki J, Dey N (2020) Texture feature extraction techniques for image recognition. Springer, Singapore
Sasank VVS, Venkateswarlu S (2021) Brain tumor classification using modified kernel based softplus extreme learning machine. Multim Tools Appl 80(9):13513–13534
Zhang M, Wang D, Yang J (2022) Hybrid-flash butterfly optimization algorithm with logistic mapping for solving the engineering constrained optimization problems. Entropy 24(4):525. https://doi.org/10.3390/e24040525
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors agreed on the content of the study. PY and SCS collected all the data for analysis. PY agreed on the methodology. PY and SCS completed the analysis based on agreed steps. Results and conclusions are discussed and written together. The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
This article does not contain any studies with human participants.
Human and animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, P., Sharma, S.C. HFBO-KSELM: Hybrid Flash Butterfly Optimization-based Kernel Softplus Extreme Learning Machine for Classification of Chronic Kidney Disease. J Supercomput 79, 17146–17169 (2023). https://doi.org/10.1007/s11227-023-05337-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-023-05337-6