Abstract
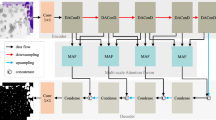
Nuclei segmentation models significantly improve the efficiency of nuclei analysis. Current deep learning models for nuclei segmentation can be divided into single-path and multi-path approaches. Single-path algorithms often underestimate the importance of edge supervision, while multi-path algorithms typically share layers but leading to potential negative impacts on feature extraction due to gradient updates during backpropagation. To address these challenges, we introduced a novel CLIP-Driven Referring model. Specifically, we designed a Class Guidance block that guides the model in distinguishing and aggregating different features by computing the similarity between images and text. We also introduced a Deformable Feature Attention block in the image branch to enhance local modeling abilities. We analyzed DICE, AJI and PQ metrics improvements through cross-dataset validation. Our model achieved increases of 4.14%, 5.69% and 9.06%, respectively, on the CPM when training with MoNuSeg, and 2.16%, 3.85% and 2.86%, respectively, on the MoNuSeg when training with CPM.





Similar content being viewed by others
Data availibility
Code is available at https://github.com/rsy980/CRNS-Net.
References
Fischer EG (2020) Nuclear morphology and the biology of cancer cells. Acta Cytol 64(6):511–519
Grünwald BT, Devisme A, Andrieux G, Vyas F, Aliar K, McCloskey CW, Macklin A, Jang GH, Denroche R, Romero JM et al (2021) Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 184(22):5577–5592
Rosellini M, Marchetti A, Mollica V, Rizzo A, Santoni M, Massari F (2023) Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat Rev Urol 20(3):133–157
Graham S, Vu QD, Raza SEA, Azam A, Tsang YW, Kwak JT, Rajpoot N (2019) Hover-net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal 58:101563
Hörst F, Rempe M, Heine L, Seibold C, Keyl J, Baldini G, Ugurel S, Siveke J, Grünwald B, Egger J et al (2024) Cellvit: vision transformers for precise cell segmentation and classification. Med Image Anal 94:103143
Zhao B, Chen X, Li Z, Yu Z, Yao S, Yan L, Wang Y, Liu Z, Liang C, Han C (2020) Triple u-net: hematoxylin-aware nuclei segmentation with progressive dense feature aggregation. Med Image Anal 65:101786
Han C, Yao H, Zhao B, Li Z, Shi Z, Wu L, Chen X, Qu J, Zhao K, Lan R et al (2022) Meta multi-task nuclei segmentation with fewer training samples. Med Image Anal 80:102481
Mongan J, Moy L, Kahn CE Jr (2020) Checklist for artificial intelligence in medical imaging (claim): a guide for authors and reviewers. Radiol Artif Intell 2(2):e200029
Dong N, Feng Q, Chang J, Mai X (2024) White blood cell classification based on a novel ensemble convolutional neural network framework. J Supercomput 80(1):249–270
Liu J, Zhang Y, Chen JN, Xiao J, Lu Y, A Landman B, Yuan Y, Yuille A, Tang Y, Zhou Z (2023) Clip-driven universal model for organ segmentation and tumor detection. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp 21152–21164
Imani M, Zehtabian A (2024) Attention based morphological guided deep learning network for neuron segmentation in electron microscopy. J Supercomput 18:1–23
Yang X, Li H, Zhou X (2006) Nuclei segmentation using marker-controlled watershed, tracking using mean-shift, and Kalman filter in time-lapse microscopy. IEEE Trans Circuits Syst I Regul Pap 53(11):2405–2414
Cheng J, Rajapakse JC et al (2008) Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans Biomed Eng 56(3):741–748
Ali S, Madabhushi A (2012) An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging 31(7):1448–1460
Liao M, Yq Zhao, Li Xh, Dai Ps Xu, Xw Zhang Jk, Bj Zou (2016) Automatic segmentation for cell images based on bottleneck detection and ellipse fitting. Neurocomputing 173:615–622
Chen H, Qi X, Yu L, Heng PA (2016) Dcan: deep contour-aware networks for accurate gland segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 2487–2496
Graham S, Rajpoot NM (2018) Sams-net: Stain-aware multi-scale network for instance-based nuclei segmentation in histology images. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, pp 590–594
Cheng Z, Qu A (2020) A fast and accurate algorithm for nuclei instance segmentation in microscopy images. IEEE Access 8:158679–158689
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M (2022) Swin-unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision, Springer, pp 205–218
Zhou Y, Onder OF, Dou Q, Tsougenis E, Chen H, Heng PA (2019) Cia-net: robust nuclei instance segmentation with contour-aware information aggregation. In: Information Processing in Medical Imaging: 26th International Conference, IPMI 2019, Hong Kong, China, June 2–7, 2019, Proceedings 26, Springer, pp 682–693
Oda H, Roth HR, Chiba K, Sokolić J, Kitasaka T, Oda M, Hinoki A, Uchida H, Schnabel JA, Mori K (2018) Besnet: boundary-enhanced segmentation of cells in histopathological images. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part II 11, Springer, pp 228–236
Jiang H, Zhang R, Zhou Y, Wang Y, Chen H (2023) Donet: Deep de-overlapping network for cytology instance segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 15641–15650
Wang Z, Lu Y, Li Q, Tao X, Guo Y, Gong M, Liu T (2022) Cris: clip-driven referring image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 11686–11695
Rao Y, Zhao W, Chen G, Tang Y, Zhu Z, Huang G, Zhou J, Lu J (2022) Denseclip: language-guided dense prediction with context-aware prompting. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 18082–18091
Wang Z, Wu Z, Agarwal D, Sun J (2022) Medclip: contrastive learning from unpaired medical images and text. In: 2022 Conference on Empirical Methods in Natural Language Processing, EMNLP 2022
Qin Z, Yi HH, Lao Q, Li K (2023) Medical image understanding with pretrained vision language models: a comprehensive study. In: The Eleventh International Conference on Learning Representations
Ye Y, Xie Y, Zhang J, Chen Z, Xia Y (2023) Uniseg: a prompt-driven universal segmentation model as well as a strong representation learner. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, pp 508–518
Shi D (2024) Transnext: robust foveal visual perception for vision transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 17773–17783
Pan X, Ye T, Xia Z, Song S, Huang G (2023) Slide-transformer: hierarchical vision transformer with local self-attention. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 2082–2091
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp 10012–10022
Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36(7):1550–1560
Vu QD, Graham S, Kurc T, To MNN, Shaban M, Qaiser T, Koohbanani NA, Khurram SA, Kalpathy-Cramer J, Zhao T et al (2019) Methods for segmentation and classification of digital microscopy tissue images. Front Bioeng Biotechnol 7:53
Kirillov A, He K, Girshick R, Rother C, Dollár P (2019) Panoptic segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 9404–9413
Naylor P, Laé M, Reyal F, Walter T (2018) Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans Med Imaging 38(2):448–459
Islam Sumon R, Bhattacharjee S, Hwang YB, Rahman H, Kim HC, Ryu WS, Kim DM, Cho NH, Choi HK (2023) Densely convolutional spatial attention network for nuclei segmentation of histological images for computational pathology. Front Oncol 13:1009681
Zhang Y, Cai L, Wang Z, Zhang Y (2024) Seine: Structure encoding and interaction network for nuclei instance segmentation. arXiv preprint arXiv:2401.09773
Lou W, Li H, Li G, Han X, Wan X (2023) Which pixel to annotate: a label-efficient nuclei segmentation framework. IEEE Trans Med Imaging 42(4):947–958
Qin J, He Y, Zhou Y, Zhao J, Ding B (2022) Reu-net: region-enhanced nuclei segmentation network. Comput Biol Med 146:105546
Deshmukh G, Susladkar O, Makwana D, Mittal S et al (2022) Feednet: a feature enhanced encoder-decoder lSTM network for nuclei instance segmentation for histopathological diagnosis. Phys Med Biol 67(19):195011
Hu Q, Chen Y, Xiao J, Sun S, Chen J, Yuille AL, Zhou Z (2023) Label-free liver tumor segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 7422–7432
Norgeot B, Quer G, Beaulieu-Jones BK, Torkamani A, Dias R, Gianfrancesco M, Arnaout R, Kohane IS, Saria S, Topol E et al (2020) Minimum information about clinical artificial intelligence modeling: the mi-claim checklist. Nat Med 26(9):1320–1324
Huynh D, Kuen J, Lin Z, Gu J, Elhamifar E (2022) Open-vocabulary instance segmentation via robust cross-modal pseudo-labeling. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 7020–7031
Acknowledgements
This work was supported by Henan Provincial Medical Science and Technology Tackling Program (LHGJ20240404), Provincial and Ministry Co-construction Key Projects of Henan Provincial Medical Science and Technology Tackling Program (SBGJ202402077).
Author information
Authors and Affiliations
Contributions
Ruosong Yuan Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft. Wenwen Zhang Data curation, Supervision, Validation, Writing—review & editing. Xiaokang Dong Writing—review & editing. Wanjun Zhang Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing—original draft, Writing— review & editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no Conflict of interest to declare that are relevant to the content of this article.
Ethical approval
No ethical approval was required for this study as it did not involve human participants or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, R., Zhang, W., Dong, X. et al. Crns: CLIP-driven referring nuclei segmentation. J Supercomput 81, 174 (2025). https://doi.org/10.1007/s11227-024-06692-8
Accepted:
Published:
DOI: https://doi.org/10.1007/s11227-024-06692-8