Abstract
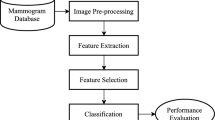
Cancer, or malignant tumor, is a group of diseases that arises from the abnormal proliferation of body cells, which have the ability to invade or spread to other parts of the body. Many researchers have proposed various methods to detect breast cancer; however, the accuracy of these methods has often been insufficient due to ineffective features selection and a lack of appropriate analytical techniques. To address this issue, we need an accurate feature extraction model. In this paper, we propose an intelligent hybrid feature extraction model for automating cancer diagnosis (IHFEACD) with high accuracy. This mathematical model generates more efficient features based on the structure of previous feature formulas. Furthermore, the proposed model combines new features with existing ones to create a new feature space for early cancer detection. Although this model can be applied to detect different types of cancer, we focus on breast cancer in women as our case study. To validate our approach, we investigated the mammographic image analysis society (MIAS) database and curated the breast imaging subset of digital database for screening mammography (CBIS-DDSM). The results indicate that the proposed method effectively classifies normal/abnormal and benign/malignant cases. By optimizing the feature structure in this new space, we have achieved improved accuracy in breast cancer diagnosis. The simulation results demonstrate high performance, showing an accuracy of 99.8%, sensitivity of 98%, and specificity of 99.4% using the naive bayes (NB) classifier on the MIAS database. Additionally, the proposed IHFEACD approach outperforms other methods in terms of accuracy metrics, achieving a 0.8 training test rate on the MIAS database, along with improvements of 0.3%, 1%, 6.8%, and 0.5% compared to IAIS-ABC-CDS, CADx, OKMT-SGO, and ANN-t-SNE approaches, respectively. For the CBIS-DDSM database, the performance results for breast cancer detection are also remarkable, with an accuracy of 99.5%, sensitivity of 98.8%, and specificity of 99.3% using both simple and naive bayes classifiers. This research provides a clearer picture of the robustness of the model across different databases. The proposed approach demonstrates significant improvements compared to previous methods from various comparative perspectives. Finally, this model has the potential to assist medical professionals in making informed decisions regarding breast cancer diagnosis.












Similar content being viewed by others
Data availability
No datasets were generated or analyzed during the current study.
References
Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, Jemal A, Siegel RL (2022) Breast cancer statistics. CA Cancer J Clin 72(6):524–541. https://doi.org/10.3322/caac.21754
Boutry J, Tissot S, Ujvari B, Capp JP, Giraudeau M, Nedelcu AM (1877) Thomas F (2022) The evolution and ecology of benign tumors. Biochim Biophys Acta Rev Cancer 1:188643
Bisoyi A (2022) Ownership, liability, patentability, and creativity issues in artificial intelligence. Info Securit Jurnal 31(4):377–386. https://doi.org/10.1080/19393555.2022.2060879
Nayak DSK, Mohapatra S, Al-Dabass D, Swarnkar T (2023) Deep learning approaches for high dimension cancer microarray data feature prediction: a review. Computational intelligence in cancer diagnosis. Elsevier, pp 13–41. https://doi.org/10.1016/B978-0-323-85240-1.00018-3
Melekoodappattu JG, Subbian PS (2023) Automated breast cancer detection using hybrid extreme learning machine classifier. J Ambient Intell Humaniz Comput 14(5):5489–5498. https://doi.org/10.1007/s12652-020-02359-3
Chaieb R, Kalti K (2018) Feature subset selection for classification of malignant and benign breast masses in digital mammography. Pattern Anal Appl 22:803–829
Srikantamurthy MM, Rallabandi VPS, Dudekula DB, Natarajan S, Park J (2023) Classification of benign and malignant subtypes of breast cancer histopathology imaging using hybrid CNN-LSTM based transfer learning. BMC Med Imag 23(1):1–15
Talukder MA, Islam MM, Uddin MA, Akhter A, Hasan KF, Moni MA (2022) Machine learning-based lung and colon cancer detection using deep feature extraction and ensemble learning. Expert System Application 205:117605
Gupta S, Agrawal S, Singh SK, Kumar S (2023) A novel transfer learning-based model for ultrasound breast cancer image classification. In: Smys S, João MR, Tavares S, Shi F (eds) Computational Vision and bio-inspired computing: proceedings of ICCVBIC 2022. Springer Nature Singapore, Singapore, pp 511–523. https://doi.org/10.1007/978-981-19-9819-5_37
Punitha S, Turjman FA, Stephan T (2021) An automated breast cancer diagnosis using feature selection and parameter optimization in ANN. Comput Electr Eng 90:106958
Ogundokun RO, Misra S, Douglas M, Damaševičius R, Maskeliūnas R (2022) Medical Internet-of-Things based breast cancer diagnosis using hyperparameter-optimized neural networks. Future Internet 14(5):153
Sharmin S, Tanvir Ahammad Md, Talukder A, Ghose P (2023) A hybrid dependable deep feature extraction and ensemble-based machine learning approach for breast cancer detection. IEEE Access 11:87694–87708. https://doi.org/10.1109/ACCESS.2023.3304628
Keshta I, Deshpande PS, Shabaz M (2023) Multi-stage biomedical feature selection extraction algorithm for cancer detection. SN Appl. https://doi.org/10.1007/s42452-023-05339-2
Kode H, Barkana BD (2024) Deep Learning- and Expert Knowledge-Based Feature Extraction and Performance Evaluation in Breast Histopathology Images. Cancers (Basel) 15(12):3075. https://doi.org/10.3390/cancers15123075
Carvalho ED, Filho AOC, Silva RRV, Araújo FHD, Diniz JOB, Silva AC, Paiva AC, Gattass M (2020) Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif Intell Med 105:101845
Chandana CH, Krishna GB (2021) Breast cancer detection using random forest classifier. Materials Today: Proceedings
Sahu Y, Tripathi A, Gupta RK (2023) A CNN-SVM based computer aided diagnosis of breast Cancer using histogram K-means segmentation technique. Multimedia Tools Applications 82:14055–14075. https://doi.org/10.1007/s11042-022-13807-x
AlShorbajit I, Kachare P, Zogaan W (2022) Learning features using an optimized artificial neural network for breast cancer diagnosis. SN COMPUT SCI 3:229. https://doi.org/10.1007/s42979-022-01129-6
Younis NK, Roumieh R, Bassil EP, Ghoubaira JA, Kobeissy F, Eid AH (2022) Nanoparticles: attractive tools to treat colorectal cancer. Seminars in Cancer Biology journal 86(2):1–13
Isosalo A, Inkinen SI, Turunen T, Ipatti PS, Reponen J, Nieminen MT (2023) Independent evaluation of a multi-view multi-task convolutional neural network breast cancer classification model using Finnish mammography screening data. Comput Biol Med 161:107023
Kavitha T, Mathai PP, Karthikeyan C (2022) Deep Learning Based Capsule Neural Network Model for Breast Cancer Diagnosis Using Mammogram Images. Interdiscip Sci Comput Life Sci 14:113–129. https://doi.org/10.1007/s12539-021-00467-y
Alickovic E, Subasi A (2020) Normalized Neural Networks for Breast Cancer Classification. In International Conference on Medical and Biological Engineering. pp. 519–524. https://doi.org/10.1007/978-3-030-17971-7-77
Singh D, Nigam R, Mittal R (2023) Information retrieval using machine learning from breast cancer diagnosis. Multimedia Tools Applicatios 82:8581–8602. https://doi.org/10.1007/s11042-022-13550-3
Chaieb R, Kalti K (2019) Feature subset selection for classification of malignant and benign breast masses in digital mammography. Pattern Anal Applic 22:803–829. https://doi.org/10.1007/s10044-018-0760-x
Gonzalez RC, Woods RE (2002) Digital image processing. Prentice- Hall Inc, New Jersey, pp 76–142
Galloway MM (1975) Texture classification using gray level run length. Computing Graph Image Process 4:172–179
Tamura H, Mori S, Yamawaki T (1978) Texture features corresponding to visual perception. IEEE Trans Syst Man Cybernet Smc 8(6):460–473. https://doi.org/10.1109/TSMC.1978.4309999
Manjunath BS, Ma WY (1996) Texture features for browsing and retrieval of large image data. IEEE Trans Pattern Anal Mach Intell (Spec Issue Digit Library) 18(8):837–842. https://doi.org/10.1109/34.531803
Rodrigues JF Jr, Traina AJM, Traina C Jr (2005) Enhanced visual evaluation of feature extractors for image mining. In: The 3rd ACS/IEEE International Conference on Computer Systems and Applications
Cheng HD, Shi XJ, Min R, Hu LM, Cai XP, Du HN (2006) Approaches for automated detection and classification of masses in mammograms. Pattern Recognit 39:646–668
Peng H, Long F, Ding C (2005) Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intelligence 27(8):1226–1238. https://doi.org/10.1109/TPAMI.2005.159
Ayadi W, Elhamzi W, Charfi I, Atri M (2019) A hybrid feature extraction approach for brain MRI classification based on Bag-of-words. Biomed Signal Process Control 48:144–152. https://doi.org/10.1016/j.bspc.2018.10.010
Dhahbi S, Barhoumi W, Zagrouba E (2015) Breast cancer diagnosis in digitized mammograms using curvelet moments. Comput Biol Med 64:79–90
Mojez H, Bidgoli AM, Javadi HHS (2023) Extended array model of star capacity-aware delay-based next controller placement problem for multiple controller failures in software-defined wide area networks. J Ambient Intell Human Comput 14:11039–11057. https://doi.org/10.1007/s12652-022-04384-w
Mojez H, Bidgoli AM, Javadi HHS (2022) Star capacity-aware latency-based next controller placement problem with considering single controller failure in software-defined wide-area networks. J Supercomput 78:13205–13244. https://doi.org/10.1007/s11227-022-04360-3
Manocha S, Girolami M (2007) An empirical analysis of the probabilistic k-nearest neighbor classifier. Pattern Recognit Lett 28:1818–1824. https://doi.org/10.1016/j.patrec.2007.05.018
Suckling J (1994) The Mammographic Image Analysis Society Digital Mammogram Database. Exerpta Medica International Congress. pp. 375–378.
Lee R, Gimenez F, Hoogi A (2017) A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data 4:170177. https://doi.org/10.1038/sdata.2017.177
Al-Tam RM, Al-Hejri AM, Narangale SM, Samee NA, Mahmoud NF, Al-masni MA, Al-antari MA (2022) A hybrid workflow of residual convolutional transformer encoder for breast cancer classification using digital X-ray mammograms. Biomedicines 10(11):2971. https://doi.org/10.3390/biomedicines10112971
Li Q (2007) Improvement of bias and generalizability for computer-aided diagnostic schemes. Computing Med Imaging Gr 31:338–345. https://doi.org/10.1016/j.compmedimag.2007.02.004
Al-Hejri AM, Al-Tam RM, Fazea M, Sable AH, Lee S, Al-antari MA (2023) ETECADx: Ensemble Self-Attention Transformer Encoder for Breast Cancer Diagnosis Using Full-Field Digital X-ray Breast Images. Diagnostics 13(1):89. https://doi.org/10.3390/diagnostics13010089
Archana R, Jeevaraj PSE (2024) Deep learning models for digital image processing: a review. Artif Intell Rev 57:11. https://doi.org/10.1007/s10462-023-10631-z
Li L, Fan Y, Tse M, Lin KY (2020) A review of applications in federated learning. Comput Ind Eng 149:106854
Acknowledgements
The authors would like to thank Dr. Hadi Mojez and Engineer Bahram Nazeri for their valuable guidance regarding MATLAB software training and simulation.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sector.
Author information
Authors and Affiliations
Contributions
All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rahmani, R., Akbarpour, S., Farzan, A. et al. A new intelligent hybrid feature extraction model for automating cancer diagnosis: a focus on breast cancer. J Supercomput 81, 651 (2025). https://doi.org/10.1007/s11227-025-07077-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s11227-025-07077-1