Abstract
Meniscal tears, a prevalent orthopedic condition caused by abrupt knee movements, excessive load, or injury, require an accurate diagnosis for effective treatment. This study investigates the vision transformer (ViT) models' efficacy in automated classification of meniscus pathologies. It also explores how feature reduction using the ElasticNet method can improve classification accuracy and computational efficiency. The study utilized MRI scans from a dataset comprising 5000 images collected from clinical cases. Initially, classification was performed using EfficientNet and SqueezeNet architectures. Subsequently, feature extraction was conducted using ViT models, generating a feature set of 1000 dimensions. ElasticNet was employed to reduce features before reclassification using support vector machines (SVM). Model performance was evaluated based on accuracy, precision, sensitivity, and specificity. The ViT_base_32 model achieved a classification accuracy of 99.9% with a processing time of 1.2 s. Feature reduction via ElasticNet significantly enhanced classification performance while maintaining high precision, sensitivity, and specificity. These improvements demonstrate the effectiveness of combining ViT models with ElasticNet to diagnose meniscal tears. The findings highlight the potential of vision transformer models, in conjunction with ElasticNet, to provide rapid and highly accurate diagnostic assistance for meniscal injuries. This methodology shows promise for application to other medical diagnostic domains, offering valuable advancements in healthcare technology.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Meniscus tears are a prevalent orthopedic condition that, if not addressed, may result in pain, diminished mobility, and chronic joint damage. Timely and precise diagnosis prevents further decline and facilitates successful therapy. Conventional diagnostic techniques, including physical examinations and MRI assessments by radiologists, depend significantly on expert analysis, which can be labor-intensive and unpredictable. Advancements in artificial intelligence have led to the emergence of deep learning-based models as potent instruments for improving diagnostic precision and efficiency in medical picture processing.
Recent research has shown substantial advancements in the automated diagnosis and categorization of meniscus tears through deep learning methodologies. Convolutional neural networks (CNNs) and vision transformers (ViTs) have been extensively utilized in medical imaging, demonstrating exceptional efficacy in feature extraction and classification applications. These algorithms examine MRI scans by identifying complex patterns within high-dimensional data, allowing for precise differentiation between normal and diseased tissues. Furthermore, deep learning algorithms enable automated diagnosis, decreasing reliance on manual assessment and lowering the probability of diagnostic inaccuracies. Consequently, these AI-driven methodologies possess the capacity to optimize healthcare workflows and enhance patient outcomes.
Magnetic resonance imaging (MRI) is the definitive method for examining soft tissues, especially in orthopedic evaluations. Because MRI images are complex, advanced computing techniques are required to improve diagnostic efficacy. Deep learning classification models have significantly expedited the diagnosis process by autonomously detecting anomalies and emphasizing crucial features that human observers may miss. As deep learning advances, its incorporation into clinical practice offers enhanced accuracy and accessibility to high-quality diagnostic tools, especially in resource-constrained healthcare environments.
2 Literature review
Medical imaging and artificial intelligence applications have significantly progressed in the last decade. In particular, several studies have addressed meniscus diagnosis using magnetic resonance imaging (MRI) to understand the effectiveness and potential of deep learning techniques. Many studies are in the literature on using deep learning approaches in medical image analysis and their role in diagnosing meniscal injuries [1, 2].
In 2018, Bien et al. created MRNet, a deep learning model to analyze knee MRI data [3]. The model was evaluated using 1,370 knee MRI scans from Stanford University to identify ACL injuries, meniscal tears, and general abnormalities. MRNet demonstrated exceptional performance with AUC values of 96.5% for ACL tears, 84.7% for meniscal tears, and 93.7% for overall abnormalities. In the external validation dataset, the model identified ACL tears with an accuracy of 82.4%, comparable to that of radiologists. Moreover, supplying the model outputs to radiologists enhanced sensitivity, particularly in diagnosing ACL rupture. The research indicated that deep learning models serve as a helpful instrument in knee MRI analysis.
Roblot et al. created a rapid regional convolutional neural network (Faster-RCNN) method 2019 to detect meniscal injuries in knee MRI images [4]. The technique was assessed utilizing a training dataset of 1123 photos and a test dataset of 700 images. The model exhibited strong performance, with an AUC of 94% for detecting meniscal tears, 92% for identifying tear location, and 83% for determining tear direction. The weighted AUC value encompassing all tasks was given as 90%. This study demonstrates that deep learning methodologies can achieve high accuracy in diagnosing meniscal tears, assisting radiologists in their diagnostic procedures.
A 2021 study by Tack and colleagues introduced a multi-task deep learning approach for detecting meniscal tears in magnetic resonance imaging (MRI) data [5]. This model, evaluated using 3D DESS and IW TSE MRI scans from the Osteoarthritis Initiative database, can identify meniscal rips distinctly in three anatomical subregions (anterior horn, body, posterior horn) of the medial meniscus (MM) and lateral meniscus (LM). A ResNet-based 3D convolutional neural network (CNN) was employed in the study, with the model performance given as AUC values of 94%, 93%, 93%, and 93% for MM, and 96%, 94%, and 91% for LM on DESS MRI scans, as well as 84%, 88%, and 86% for MM, and 95%, 91%, and 90% for LM on IW TSE scans. This approach offered anatomically precise localization of meniscal tears and was intended to be readily applicable to various MRI sequences.
Rizk and colleagues created a deep learning algorithm in 2021 to detect and characterize meniscal tears in adult knee MRI data [6]. The research used an extensive dataset comprising 11,353 knee MRI scans from 10,401 people. The AUC values for detecting medial and lateral meniscal tears utilizing 3D convolutional neural networks (CNN) are 93% and 84%, respectively. In comparison, the identification of meniscal tear migration yields AUC values of 91% and 95%, respectively. The model underwent external validation using the MRNet dataset, resulting in AUC values elevated to 89% following fine-tuning. The research illustrated the precision of deep learning algorithms in diagnosing meniscal tears and their capacity to aid radiologists in practical settings.
In a 2022 study by Jie Li and associates, a deep learning model was employed to detect and diagnose meniscal tears using magnetic resonance imaging (MRI) [7]. A network architecture utilizing the Mask RCNN model and ResNet50 was constructed in the study. The dataset utilized in the study comprises MRI pictures from 924 individuals employed in the training, validation, and testing phases. The model's efficacy was additionally corroborated using external datasets and arthroscopic surgery. The findings indicated that the model can accurately differentiate between healthy, torn, and degenerative menisci. The diagnostic accuracy was documented as 87.50% for intact meniscus, 86.96% for ruptured meniscus, and 84.78% for degenerative meniscus. The model identified torn and intact meniscal tears with over 80% accuracy using 3.0 Tesla MRI images.
In a 2023 study by Harman and colleagues, a deep learning model was constructed for detecting meniscal tears using rapid magnetic resonance imaging (MRI) data [8]. The research was conducted using the FastMRI + dataset and employed three distinct deep learning-based image reconstruction models: U-NET, iRIM, and E2E VarNET. The nnDetection model was incorporated to detect meniscal tears, and the efficacy of these models was assessed on both 4 × and 8 × accelerated MRI images. E2E VarNET had superior average accuracy ratings, achieving 69% with a fourfold speedup and 67% with an eightfold speedup. The research indicated that deep learning-based reconstruction techniques are excellent in maintaining and identifying clinically significant pathology information.
A 2024 study by Botnari and associates thoroughly assessed the efficacy of deep learning models on knee MRI images for diagnosing and classifying meniscal injuries [9]. This comprehensive review and meta-analysis demonstrated that multiple deep learning models could accurately identify medial and lateral meniscal tears, as well as tears in the anterior and posterior horn, with AUC values between 83 and 94%. The study underscores the capability of deep learning techniques to identify and categorize meniscal tears, occasionally surpassing the efficacy of radiologists. The models exhibited a sensitivity of 83% and a specificity of 85%. The research examines the difficulties and novel solutions provided by deep learning models in diagnosing intricate medical diseases, including meniscal tears.
Contemporary deep learning models for meniscal tear diagnosis exhibit great accuracy rates but have significant limitations. Current research frequently utilizes extensive and homogeneous datasets, which inadequately represent the variability present in clinical situations. The MRNet model, created by Bien et al., was evaluated using a singular dataset from Stanford University, raising concerns about its generalizability to MRI scans from various imaging systems. Likewise, the Faster-RCNN model introduced by Roblot et al. was assessed on a restricted set of pictures, and it remains uncertain if the model can sustain equivalent accuracy in broader patient cohorts. Many research have generalization challenges owing to insufficient data diversity, which restricts their direct applicability in clinical settings.
Moreover, although CNN-based models (e.g., ResNet, DenseNet), prevalent in current literature, are proficient in deep feature extraction, they are constrained by substantial computational expenses and extended processing durations. These models are particularly deficient in properly processing high-dimensional features in extensive MRI datasets. Although the model created by Rizk et al. was evaluated on an extensive patient dataset, it fails to deliver a definitive evaluation of the model’s processing time and therapeutic relevance. Furthermore, the majority of research concentrate solely on picture classification, neglecting to incorporate supplementary clinical data (e.g., patient history, physical examination results) to enhance the medical diagnostic process. The opaque nature of deep learning models is a significant barrier, as the interpretability of judgments made by most models is restricted and lacks explainable frameworks that would enhance reliability in clinical applications. In light of these deficiencies, our present study seeks to address the gaps in the literature by providing diminished processing time, enhanced generalizability, and superior classification accuracy through the integration of vision transformer (ViT) and ElasticNet.
Research on the efficacy of vision transformer (ViT) models in medical picture categorization is scarce, with prevailing studies predominantly emphasizing traditional deep learning techniques that necessitate extensive datasets. Moreover, these approaches frequently exhibit inefficiency regarding processing times and have been evaluated on a restricted dataset of actual patient information. Our objective is to address these deficiencies.
-
This work revealed that ViT surpasses current deep learning methodologies by achieving high accuracy (99.9%) and advanced feature extraction capabilities in diagnosing meniscal tears, given the scarcity of ViT research in the literature.
-
This study addressed the challenges of processing time and resource efficiency, which are hardly discussed in the literature. It optimized the extensive array of retrieved characteristics from ViT using ElasticNet. This method considerably diminished the processing duration (from 3387 s to 3.56 s) while enhancing the classification accuracy to 99.9%.
-
This work evaluates the model utilizing a comprehensive collection of MR images derived from actual patient data, contributing a novel dataset to the existing literature, in contrast to many studies that rely on generic or synthetic datasets.
-
Integrating deep learning and machine learning techniques commonly employed independently in the literature, this hybrid modeling strategy has enhanced meniscus diagnosis accuracy rates and showcased these technologies' synergistic potential.
-
Our research has addressed the efficiency challenges commonly faced in medical image classification. By reducing features and optimizing the models, we have produced faster and more efficient classification models with fewer features.
Existing research shows that deep learning models for meniscal tear diagnosis offer high accuracy but have limitations such as generalizability, interpretability, and processing time. The proposed ViT + ElasticNet + SVM method aims to address these shortcomings with 99.9% accuracy and low processing time (0.12 s). ElasticNet improves the processing time by eliminating redundant features, while SVM supports the model with its strong classification capacity. This combination is expected to provide a faster and more reliable diagnosis process, making deep learning-based systems more suitable for clinical use.
3 Methods
3.1 Dataset
The precise diagnosis of meniscus tears is accomplished with magnetic resonance imaging (MRI). MRI is a noninvasive, radiation-free modality that provides high-resolution imaging of soft tissues, facilitating comprehensive evaluation of meniscus tears [10,11,12,13]. Various MRI sequences enable the assessment of the kind, size, and connection of tears with adjacent tissues. The assessment of MRI images relies on the expertise of radiologists, making the process time-consuming and subject to inter-observer variability. Consequently, computerized analysis methods based on deep learning are increasingly favored to expedite diagnostic procedures and improve accuracy [1, 2, 14].
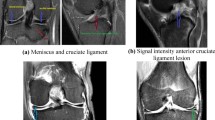
This study utilizes a dataset including 1499 meniscus MRI images, exclusively sourced from Fırat University Hospital and employed for the first time here. The dataset images are categorized into two primary classes to aid in the classification of meniscus tears: healthy and tear. Expert radiologists assessed the images, excluding MRI scans of low diagnostic quality or those with imaging artifacts from the study. During the dataset creation, only images exhibiting distinctly defined tears and high resolution were chosen, guaranteeing that the model was trained using a dependable data source [15, 16].
To improve dataset accuracy and assure classification reliability, the criteria for including tear images have been specified. The classification was based on the location of the tears within the meniscus, their kind (e.g., radial, horizontal, complicated), and their association with the bone. The labeling process was conducted separately by two professional radiologists, each with a minimum of 10 years of experience, and checked for accuracy by the consensus technique [1, 2]. Furthermore, to preserve image quality, all MRI scans were conducted utilizing machines with a consistent slice thickness and identical magnetic field strength.
Cross-validation has been employed to mitigate overfitting and enhance the model's generalization ability. Specifically, the fivefold cross-validation method was used to assess the model's performance across different data subsets and ensure its robustness [10,11,12,13]. This approach allowed the model to be trained and validated on multiple partitions of the dataset, reducing bias and improving its ability to generalize to unseen data. Additionally, data augmentation techniques, including rotation, brightness modification, and contrast adjustment, were applied to further enhance model robustness [14]. Figure 1 shows representative images from the dataset utilized in the study.
This study's dataset underwent particular preprocessing procedures to enhance model performance and guarantee reproducibility. Initially, all photos were downsized to 224 × 224 pixels to conform to the input dimensions of the vision transformer (ViT) models. Subsequently, data augmentation approaches were employed to enhance data diversity and expand the model's generalization capability. These methods encompass rotation, horizontal translation, and random cropping. Several transformations were implemented to prepare the photos for input: The images were scaled (transforms.Resize), converted to tensor format (transforms.ToTensor), and normalized (transforms.Normalize) using the specified transforms. Construct a function. In the normalization step, the mean values ([0.485, 0.456, 0.406]) for each channel of each image were obtained and scaled by the standard deviation ([0.229, 0.224, 0.225]). These procedures were executed to facilitate accelerated learning for the model and to represent the properties of the input data.
3.2 Feature extraction algorithms
The vision transformer architecture, first introduced by Google Brain in 2020 and successful in natural language processing, has started to be used in image processing due to this success. This architecture first examines the relationships between languages and then uses the exact mechanism to analyze images [17]. Studies have shown effective results, especially in image classification [18].
ViT divides image data into fixed-size patches and processes them like words in natural language processing [19]. Each patch is converted into a set of numeric vectors, considered input, and combined with a position encoding.
Transformer blocks utilize a multi-head self-attention technique that facilitates the assessment of significance among patches [18]. This method facilitates the more efficient analysis of images by concurrently processing various patch combinations. The ViT models comprise vit_base, vit_large, and vit_huge. Vit_base comprises 12 layers and 86 million parameters; vit_large consists of 24 layers and 307 million parameters, while vit_huge contains 32 layers and 632 million parameters. Vision transformers (ViT) excel on large-scale datasets, attaining comparable or superior outcomes to traditional CNNs on datasets like ImageNet. This achievement is attributable to ViT's ability to comprehend visual features in a broader context. Nonetheless, owing to the substantial data and processing resource demands, transfer learning is advised for small datasets. ViT can transform fields such as image classification, object recognition, and picture segmentation, representing a significant advancement in deep learning research.
ElasticNet is a regularization technique for linear regression models used in statistical and machine learning contexts. It combines Lasso and Ridge regression techniques, offering the advantages of both methods [20]. This approach helps to increase the model’s generalizability, especially in high-dimensional datasets and the presence of interrelated (multi-linked) features. ElasticNet adds a penalty term to optimize the regression coefficients; this penalty term includes both the L1 norm (Lasso) and the L2 norm (Ridge) [21]. In this way, both feature selection and overfitting can be avoided by controlling the complexity of the model.
The mathematical model of ElasticNet is expressed as in Eq. 1:
In Eq. 1, y denotes the dependent variables, X signifies the matrix of independent factors, and β indicates the regression coefficients [22]. λ1 and λ2 denote the L1 and L2 regularization parameters, respectively. The L1 term facilitates feature selection, whereas the L2 term enhances stability. These characteristics enable ElasticNet to surpass Lasso in datasets with correlated features. ElasticNet is extensively utilized in domains such as bioinformatics, genetics, and finance, enhancing model accuracy and generalizability by reducing duplicated features. This approach is frequently favored in data analysis and machine learning applications for offering effective answers to modeling difficulties.
The regularization parameters λ1 and λ2 in the ElasticNet model are critical in optimizing the model's performance. In this study, the values of λ1 and λ2 were optimized using k-fold cross-validation to effectively balance the feature structure of the dataset and the model's performance. λ1 aids in eliminating irrelevant and redundant features, while λ2 maintains the weight balance among correlated features, enhancing the model's stability. This regularization prevented overfitting and optimized the classification accuracy and processing time by selecting the most meaningful features.
The impact of parameter settings was evaluated by analyzing the variations in model accuracy, precision, sensitivity, and processing time under different λ1 and λ2 values. These analyses demonstrated that selecting optimal values for λ1 and λ2 enabled the efficient utilization of high-dimensional features extracted by ViT models during the classification process. The results highlight that appropriate tuning of the regularization parameters significantly improves the model's performance and generalizability.
3.3 Proposed method
Deep learning and machine learning techniques have significantly advanced medical image classification, particularly in the detection of meniscal injuries. However, existing methods still face challenges in terms of computational efficiency, generalizability, and interpretability. Traditional deep learning models, such as convolutional neural networks (CNNs), require extensive labeled data and substantial computational resources to train effectively. Moreover, CNN-based approaches often suffer from high-dimensional feature spaces, leading to redundancy and inefficiencies in processing large-scale medical imaging datasets [23,24,25]. On the other hand, classical machine learning algorithms, such as support vector machines (SVM), have demonstrated strong performance in medical image classification but are limited by their reliance on handcrafted features and their inability to leverage the rich hierarchical representations learned by deep networks [20, 26].
To address these limitations, this study proposes a hybrid framework that integrates vision transformer (ViT) models with ElasticNet and SVM for meniscal tear classification. Vision transformers have emerged as a powerful alternative to CNNs, leveraging self-attention mechanisms to extract global contextual information from images efficiently [27,28,29,30,31]. However, ViT models typically require large datasets and suffer from excessive computational complexity. By incorporating ElasticNet for feature selection, our method optimizes the deep features extracted from ViT, reducing the dimensionality while preserving the most informative representations. This approach significantly enhances classification accuracy while minimizing processing time, making it more suitable for real-time clinical applications.
The proposed methodology follows a multi-step approach. First, meniscal MRI images are processed using EfficientNet and SqueezeNet architectures to extract feature maps, as shown in Fig. 2. EfficientNet optimizes model scaling across depth, width, and resolution dimensions, whereas SqueezeNet achieves competitive accuracy with significantly fewer parameters, making it ideal for memory-constrained environments [32,33,34]. These extracted feature maps are then refined using ViT models, which apply self-attention mechanisms to learn spatial dependencies and extract high-level features. By leveraging the ElasticNet regularization technique, the method eliminates redundant and less significant features, ensuring that only the most relevant attributes contribute to the final classification step [35].
In the final stage, SVM is employed as the classifier, utilizing the optimized feature set to distinguish between healthy and torn menisci. SVM has proved highly effective in high-dimensional medical imaging tasks, as it identifies an optimal decision boundary that maximizes the margin between different classes [13, 36, 37]. The hyperplane selection process ensures robustness in classification, even with limited training data, making SVM a suitable choice for clinical applications where data variability is a concern. The integration of ViT and SVM bridges the gap between deep feature extraction and classical machine learning classification, offering a balance between interpretability, efficiency, and accuracy (Table 1).
ViT models are deep learning algorithms that successfully process visual data. For this reason, they are frequently used in classification studies. The proposed method uses ViT models for feature extraction instead of classification. With this different approach, features are extracted from the intermediate layers of the model, and these features are then reduced using ElasticNet. The reduced features are then input to the SVM machine learning algorithm.
This approach utilizes the powerful feature extraction capabilities of the ViT model to improve the classification performance of classical machine learning models. The proposed method shows that the highly significant features extracted by the ViT model can be better processed by machine learning algorithms, achieving higher accuracy rates. The flow diagram of the proposed method is given in Fig. 3.
The pseudo-code of the study is given in Table 2.
The use of ViT models for feature extraction and classification is a relatively new approach compared to deep learning models. Studies have obtained successful results. ViT is a deep learning model that allows images to be processed in parts and the relationships between these parts to be learned. Unlike traditional deep learning models, feature extraction using ViT is based on the principle of breaking the image into small pieces and applying transformer mechanisms to these pieces.
In this study, ViT models are used for feature extraction. Figure 4 summarizes feature extraction with ViT. In the first step of feature extraction, the image is processed appropriately. This step is important to provide data in the appropriate format and quality for the model's input. ViT models are designed to work with fixed-size images. Therefore, they are usually trained with images of 224 × 224 pixels. Normalization of the input data is also important for the model to perform better and learn faster. Normalization is the process of scaling each pixel value using a specific mean and standard deviation. Since deep learning models are usually processed in batches, adding the batch size to process a single image is necessary.
After feature extraction, ViT models divide the images into fixed-size patches and work on these patches. Each patch is used as an input. Each patch becomes a flattened vector and is transformed into embedding vectors by applying a linear transformation to fit the input of the model. Flattening and transforming the image segments into embedding vectors is an important step. This step brings each image segment into a form that the model can understand. The flattened segments are transformed into embedding vectors before being used as input to the ViT model. This transformation is performed using a linear layer (fully connected layer). Transformer models use positional coding to preserve positional information in sequential data. Positional coding is added to the image segments so that the model can learn the position of each segment in the image.
In ViT models, positional coding is used to preserve the positional information of sequential data. Positional coding helps the model to understand the position of each part. Thus, the model pays attention to the content and the sequence order of the parts. The next step of the process is one of the key features of the transformer architecture. This step is necessary to ensure that the ViT model works efficiently and accurately. Another fundamental component of the model is the transformer layers. These layers use attention mechanisms to learn the relationships between image parts. A transformer layer consists of multi-head attention, layer normalization, feed-forward layer, and residual connections.
The final step in obtaining the final feature vector is combining the outputs from the image segments processed by the transformer layers. This step generates the summary information necessary for the task that the model layers will use. It describes in detail how to perform feature extraction for medical imaging problems such as meniscus tear diagnosis using ViT. This process plays an important role in medical diagnostics by providing high accuracy rates.
The proposed method of the study utilizes the capabilities of vision transformer (ViT) models to provide an efficient feature extraction and classification process for meniscal tear detection. In particular, using four different ViT models, ViT_b_16, ViT_b_32, ViT_l_16, and ViT_l_32, 1000 features are extracted for each model. These features reflect the capacity of ViT models to extract rich and deep information, while the use of multiple ViT models aims to assess the ability of each model to capture details at different scales and resolutions. These extracted features form a large feature set that better represents the complexity and diversity of the dataset, which contributes to the high accuracy of the models.
The 1000 extracted features are then reduced with ElasticNet. ElasticNet combines both L1 and L2 regularization methods to increase the model’s generalizability while minimizing the impact of redundant or overly informative features. In this process, features are reduced to 298 for the ViT_b_16 model, 315 for the ViT_b_32 model, 278 for the ViT_l_16 model, and 272 for the ViT_l_32 model. This reduction process identifies the most meaningful and effective features for each model, allowing the model to perform better with less complexity. As a result, the SVM classifier is trained using the reduced features, and this approach allows for superior results in accuracy and efficiency in diagnosing meniscal tears.
This study favors vision transformer (ViT) models for their profound feature extraction capabilities and exceptional performance in identifying intricate patterns in medical images. In contrast to traditional convolutional neural networks, ViT models meticulously examine each segment of images by partitioning them, hence yielding a more comprehensive feature set. The ElasticNet approach was used for feature reduction as it integrates both L1 and L2 norms to balance high-dimensional and interdependent information. This strategy effectively minimizes duplicate or excessive features while retaining the essential ones. Alternative techniques for feature reduction, including principal component analysis (PCA) and recursive feature elimination (RFE), were also assessed. Nonetheless, PCA compromises the original significance of the characteristics, whereas RFE is less favored because of its greater processing demands. The balanced and computationally efficient framework of ElasticNet demonstrated enhanced performance consistent with the study's accuracy and processing time objectives.
This study employs CNN-based models (EfficientNet and SqueezeNet) and vision transformer (ViT) models for distinct objectives. The EfficientNet and SqueezeNet models were utilized to assess classification performance and establish a benchmark for ViT models. These models were selected for their minimal computational expense and benchmark efficacy. The study focused on ViT models because of their profound feature extraction capabilities and superior accuracy on extensive datasets. The characteristics derived from ViT models were refined by ElasticNet, leading to enhanced efficiency and precision in classification tasks. The approaches were directly compared, and the performance disparities between ViT and CNN-based models were investigated comprehensively. In summary, CNN-based models serve a supplementary function within the study’s overall methodology, whereas ViT models seek to deliver an enhanced solution for diagnosing meniscal tears.
4 Experimental results
In the study conducted in this paper, successful results were obtained with the proposed approach to meniscal tear classification. The study’s main objective is to look at the contribution of ViT models to classification studies with machine learning models by using ViT models for feature extraction and then reducing the selected features with ElasticNet. The proposed method shows that using ViT models for feature extraction and ElasticNet for feature reduction positively affects the classification results. The meniscus dataset used in the study was divided into two different classes: healthy and meniscus tear, and the studies were carried out in this context.
4.1 Traditional classification results
The study examined the traditional classification results of ViT models and CNN architectures EffNet and SqueezeNet models.
Table 3 compares the classification accuracy rates and processing times of CNN-based EffNet and SqueezeNet models used to diagnose meniscal tears. As seen in the table, the EffNet_b1 model showed the highest performance with a 95.33% accuracy rate and achieved a faster result than other EfficientNet variants with a processing time of 676.2 s. EffNet_b2 and EffNet_b3 models showed similar performance with 93% and 94.33% accuracy rates, respectively, while their processing times were 649.8 and 799.8 s. EffNet_v2_s and EffNet_v2_m models have 94% and 93% accuracy rates, while EffNet_v2_m has a longer processing time of 1050 s. EffNet_v2_1 has the lowest performance with 91.66% accuracy and the longest processing time of 1800s. SqueezeNet1_0 performs very efficiently, with an accuracy of 92.66% and a processing time of 600 s. It is faster than the other models, especially regarding processing time. These results show that SqueezeNet can be a fast and efficient option, but EffNet_b1 is superior in high accuracy rates.
Figure 5 shows the confusion matrix of the EffNet_b1 model with the highest accuracy and the EffNet_b2 model with the lowest time.
The confusion matrix indicates that the EffNet_b1 model achieves the maximum accuracy, with 200 true positives (TP) and 1 false positive (FP) for the healthy class. In the tear class, there were 86 true negatives (TN) and 13 false negatives (FN) recorded. This indicates that the model effectively differentiates healthy persons from those with tears. The EffNet_b2 model demonstrates reduced accuracy, yet it possesses a shorter runtime. This model achieved 192 true positives (TP) and 9 false positives (FP) for the healthy class, as well as 87 true negatives (TN) and 12 false negatives (FN) for the torn class. EffNet_b2 may excel in resource-limited settings due to its reduced computing time, yet EffNet_b1 demonstrates higher accuracy. The results indicate that both models possess distinct advantages, although their preferences may vary based on the specific application context. The ROC curves of the studies are shown in Fig. 6.
The study’s success is measured through performance evaluation metrics based on the complexity matrix. Recall, precision, and F1-score are used as performance evaluation metrics. Accuracy is the ratio of correctly predicted values to the total test data [38]. It characterizes the model by giving an idea of whether it works correctly. The calculation required to calculate the accuracy measure is as given in Eq. 2.
Precision value refers to the accuracy of model predictions [38]. It is the calculation of the proportion of the value that the model predicts as positive over the test data that is positive. The calculation required to calculate the precision measure is performed as shown in Eq. 3.
Recall is a frequently used performance measurement method, primarily when defining the model as a classification problem [38]. This criterion calculates the rate at which the class values that should be predicted as positive are selected as positive. If the recall measure is high, it is inferred that the model sensitivity is also high. Eq. (4) gives the necessary operations for calculating the recall measure.
F1-score is a performance measurement method obtained by taking the harmonic average of precision and recall performance measures [38]. This method aims to control the extremes in model performance. Eq. (5) gives the necessary operations for calculating the accuracy measure.
Table 4 compares the success metrics of the EffNet_b1 and EffNet_b2 models. With a recall of 94.12%, the EffNet_b2 model detects positive samples slightly better than the EffNet_b1 model, which has a recall of 93.90%. However, EffNet_b1 outperforms EffNet_b2 in precision and overall balance, with 99.50% precision and 96.62% F1-score.
In the other part of the study, traditional classification was performed with ViT architectures to compare with the results of CNN models. The results and training times are shown in Table 5.
Table 5 shows the classification results using different ViT models. The ViT_base_16 model performed the best with 97.7% accuracy but took the longest time with a processing time of 2717.52 s. The ViT_base_32 model has an accuracy of 95.32% and runs the fastest with a processing time of 1052.62 s. The ViT_large_16 model performs well with an accuracy of 97.32% but has the longest processing time of 7704.94 s. The ViT_large_32 model offers a balanced performance with an accuracy of 96.7% and a processing time of 2076.45 s. These results show that ViT models offer high accuracy rates, but the processing times vary significantly depending on the model size and configuration.
Figure 7 shows the confusion matrices of the ViT_base_16 model, which has the highest accuracy rate among ViT models, and the ViT_base_32 model, which has the shortest training time.
The confusion matrices indicate that the ViT_base_16 model exhibits the maximum accuracy, although the ViT_base_32 model is notable for its reduced training duration. The ViT_base_16 model records 82 true positives (TP) and 5 false positives (FP) for the healthy class, alongside 210 true negatives (TN) and merely 2 false negatives (FN) for the tear class. This indicates that the model operates highly sensitively and can effectively differentiate between classes. Conversely, the ViT_base_32 model exhibited a marginal decline in accuracy despite a reduced training duration, recording 75 true positives, 12 false positives, 87 true negatives, and 12 false negatives. The results indicate that the ViT_base_16 model excels in accuracy. However, the ViT_base_32 model is more advantageous in low-resource settings due to its expedited processing times. The equilibrium between these two models necessitates a selection based on the intended use and context. The ROC curves of the studies are shown in Fig. 8.
Table 6 compares the success metrics of the ViT_base_16 and ViT_base_32 models. The ViT_base_16 model has a higher F1-score (95.91%) with 97.62% recall and 94.25% precision than the ViT_base_32 model with 97.4% recall and 86.21% precision. These results show that the ViT_base_16 model offers a more balanced and superior performance in correctly identifying the positive class (recall) and making accurate optimistic predictions (precision).
4.2 Classification results of feature extraction with ViTs
In another part of the study, SVM machine learning model is used for classification. In this section, ViT models are used for feature extraction. These features are given as input to the machine learning model, and classification is performed. Table 7 shows the accuracy of the classification study performed by giving the features obtained from ViT models as input to the machine learning model. When Table 7 is analyzed, the most successful accuracy rate is 93.33% with vit_large_32.
Table 7 compares the training times of the classification runs with different ViT models. The ViT_base_16 model has the longest training time of 2640 s, indicating that it may involve more complex calculations or more parameters. In contrast, the ViT_base_32 model has the shortest training time of 1080 s, and this time was recorded as 1200 s for the ViT_large_16 and ViT_large_32 models, showing that both models perform similarly in terms of training times. These results suggest that the ViT_base_32 model can save time by providing faster training, but there is a need to balance accuracy performance with model size.
4.3 Classification results with feature extraction and feature selection
Up to this point in the study, 1000 features were selected for each model in the feature selection made with ViT. In the rest of the study, ElasticNet was used for feature reduction (Table 8).
In the continuation of the study, ResNet18, ResNet50, and ResNet101 models were also included to elaborate the study. After feature extraction with ResNet models, ElasticNet model was used to select the most meaningful features. The most significant features were classified with different machine learning models, and the results were compared. Table 9 shows the results obtained from the study.
Table 9 presents the classification results with different machine learning algorithms after the features extracted with ResNet models are optimized with the ElasticNet method. The ResNet50 model generally achieved the highest accuracy rates, particularly 94.35% accuracy with XGBoost and logistic regression. The processing time of these models is also relatively low, taking only 0.07 s with logistic regression. ResNet101 achieved the best result with the XGBoost model with 94.69% accuracy, although in this case, the processing time was slightly higher compared to ResNet50. ResNet18, on the other hand, lagged behind the other models in terms of accuracy but offered a fast processing time with logistic regression, which is advantageous in situations where low computational resources are needed.
Among all the ResNet models, logistic regression showed a balanced performance in terms of both processing time and accuracy. However, XGBoost and random forest algorithms also achieved high accuracy rates but required higher processing times. More complex algorithms, such as gradient boosting and CatBoost, did not provide a clear advantage in terms of accuracy, but their processing times were considerably longer. The results show that different models can be preferred in terms of accuracy and processing time depending on the feature structure of the dataset and the application purpose. In particular, the combination of ResNet50 and logistic regression offers an ideal balance for applications that require fast and accurate results.
In the other part of the study, features were extracted using EffNet models. The most meaningful features were selected with ElasticNet, and classification was performed using different machine learning models. Table 10 shows the results obtained from the study.
Table 10 shows the classification performance with different machine learning algorithms after the features extracted with EffNet models are optimized with ElasticNet. The EffNet_b1 model achieved the highest overall accuracy (94.35%) and the lowest processing time (0.15 s). In particular, SVM and logistic regression algorithms provided this model with high accuracy and short processing times. More complex algorithms such as XGBoost and CatBoost also achieved relatively high accuracy rates but with longer processing times. Among the other EffNet versions, the b2 model stood out with an accuracy of 92.36% and the v2_l model with an accuracy of 90.36%. However, these models did not achieve the overall stability of the b1 model.
The accuracy rates of the EffNet_v2 models are generally lower. For example, the v2_s model achieved 87.38% accuracy with XGBoost and 89.70% with logistic regression. Complex algorithms such as gradient boosting and CatBoosting were characterized by long processing times when run with v2 models. These results show that the EfficientNet_b1 model, after being optimized with ElasticNet, provides a balanced and efficient performance in terms of both accuracy and processing time. Among the machine learning algorithms, SVM and logistic regression provided the fastest and most accurate results in general. At the same time, gradient boosting and CatBoost were notable for their long processing times, although they did not show a clear advantage in accuracy. This emphasizes that the choice of model and algorithm should be based on the application context and resource constraints.
In the last stage of the study, the most significant features extracted with the proposed ViT models are selected with ElasticNet. The most significant features are classified with different machine learning models. Table 11 shows the results obtained from the study.
Table 11 presents the classification results with different machine learning algorithms after the features extracted with ViT models were optimized with ElasticNet. The ViT_b_32 model performed the best, with an accuracy of 99.9% and a processing time of only 0.12 s. In particular, the SVM algorithm outperformed all other models with this model. The ViT_b_16 model performed similarly and ranked second with 99.7% accuracy. The logistic regression and LightGBM algorithms generally provided effective results with low processing times but did not achieve as high accuracy rates as SVM. On the other hand, more complex algorithms, such as random forest and gradient boosting, had higher processing times and accuracy rates of around 93–94%.
The results in Table 11 show that optimizing the features extracted by ViT models with ElasticNet is an effective classification strategy. While the ViT_l_16 and ViT_l_32 models generally exhibited lower accuracy rates, they still performed above 90%. Among all models, the combination of ViT_b_32 and SVM provided the most balanced and efficient results in terms of both accuracy and processing time. These results show that ViT models have significant potential in medical image classification with high accuracy rates and low processing times, mainly when applied to feature selection with ElasticNet. Moreover, considering that SVM gives better results than other machine learning algorithms, it can be said that this algorithm is a suitable choice for sensitive classification tasks such as medical imaging.
Table 12 shows the number of features selected when reducing the 1000 features extracted from the ViT models using ElasticNet in SVM, which is the most successful result. For the ViT_base_16 model, 298 features were selected, indicating that the model provides an efficient representation while preserving detailed and important information. For the ViT_base_32 model, 315 features were selected, preserving more features than the other models, indicating that the model works with a broader range of information. For the ViT_large_16 and ViT_large_32 models, 275 and 272 features were selected, respectively, reflecting a feature selection strategy to retain less but more important information. This reduction process contributes to improving the performance of the models by eliminating redundant data, thus reducing the computational burden and improving model accuracy and generalizability.
Table 12 shows the classification results of the features extracted from the ElasticNet-reduced ViT models using the SVM algorithm. Recall values are 99.9% for all models, indicating that each model successfully detects positive examples. The ViT_base_32 model achieved the highest result regarding correct positive predictions and overall performance, with 99.9% precision and F1-score. The ViT_base_16 model has a balanced performance with 98.99% precision and 99.49% F1-score. The ViT_large_16 and ViT_large_32 models have a precision of 98% and an F1-score of 98.99%, slightly lower than the other models but still showing a high performance. These results reveal that ElasticNet-reduced features are effectively classified with SVM, and all models generally offer high accuracy rates. The confusion matrices of the models in the study are given in Fig. 9.
The ROC curves of the studies are shown in Fig. 10.
ViT_base_32 achieves the best accuracy (99.9%) and the quickest processing time (1.2 s) of its better-balanced design regarding size and structure relative to other ViT versions. ViT_base_32 efficiently performs deep feature extraction with fewer layers and parameters, minimizing processing time. This enables the model to rapidly and efficiently acquire significant features from intricate medical imaging data. The feature reduction executed with ElasticNet guarantees that the model utilizes the most significant features, enhancing classification accuracy and minimizing redundant processing overhead. The coherent architecture distinguishes the ViT_base_32 model from other variants in terms of accuracy and efficiency. This device offers an optimum approach for diagnosing meniscal tears in clinical applications.
5 Discussion and conclusion
This research investigated the effectiveness of vision transformer (ViT) models in diagnosing meniscal tears and their capacity to optimize classification performance when integrated with ElasticNet. ViT models have demonstrated their deep feature extraction capabilities in complex medical images such as meniscal tears. Experiments with various ViT models show that each model achieves high accuracy, recall, and precision rates. In particular, the ViT_base_32 model outperformed the other models in terms of classification accuracy (99.9%) and processing time (1.2 s). ElasticNet helped to eliminate duplicate or redundant information by selecting only the most meaningful features in the feature reduction process, thus optimizing the performance of the models. This combination provides a new standard in medical image classification [39,40,41].
However, the study has some limitations. The dataset used is only from a single institution, providing a limited context for the generalizability of the model. The generalizability of the model to different populations and imaging devices needs to be improved. In addition, the high computational power requirement of ViT models poses a significant challenge for low-resource clinical settings. This may reduce practical applicability, especially when resources are limited. Furthermore, the study did not test the performance of the proposed method in real clinical scenarios. More data on the sensitivity and reliability of the model in real-time diagnostic scenarios are required.
The findings of the study show that the combination of ViT and ElasticNet offers significant advantages compared to existing CNN-based models. However, the computational requirements of ViT may be a disadvantage in cases where CNN-based models can provide similar accuracy rates with fewer resources. Given that CNN-based models such as ResNet and DenseNet are more common in the literature in terms of generalizability, ViT models need to be tested on larger datasets to improve their generalizability.
Three main areas for future work are suggested. First, it should aim to increase the generalizability of the model with larger and more diverse datasets. Second, optimized algorithms should be developed to reduce the computational demands of ViT models. Third, prospective studies should be conducted to evaluate the effectiveness of the model in real clinical settings, and the applicability of the proposed method to other medical imaging problems should be investigated. For example, the impact of ViT on different orthopedic problems or multiple diagnostic procedures could be studied. These improvements will increase both the scientific and clinical applicability of the study.
Data availability
The data supporting this study's findings are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
Luvsannyam E, Jain MS, Leitao AR, Maikawa N, Leitao AE (2022) Meniscus tear: pathology, incidence, and management. Cureus. https://doi.org/10.7759/cureus.25121
Bhan K (2020) Meniscal tears: current understanding, diagnosis, and management. Cureus. https://doi.org/10.7759/cureus.8590
Bien N et al (2018) Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med 15(11):e1002699. https://doi.org/10.1371/journal.pmed.1002699
Roblot V et al (2019) Artificial intelligence to diagnose meniscus tears on MRI. Diagn Interv Imaging 100(4):243–249. https://doi.org/10.1016/j.diii.2019.02.007
Tack A, Shestakov A, Lüdke D, Zachow S (2021) a multi-task deep learning method for detection of meniscal tears in MRI data from the osteoarthritis initiative database. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2021.747217
Rizk B et al (2021) Meniscal lesion detection and characterization in adult knee MRI: a deep learning model approach with external validation. Physica Med 83:64–71. https://doi.org/10.1016/j.ejmp.2021.02.010
Li J et al (2022) Identification and diagnosis of meniscus tear by magnetic resonance imaging using a deep learning model. J Orthopaedic Translat 34:91–101. https://doi.org/10.1016/j.jot.2022.05.006
Harman F, Selver MA, Baris MM, Canturk A, Oksuz I (2023) Deep learning-based meniscus tear detection from accelerated MRI. IEEE Access 11:144349–144363. https://doi.org/10.1109/access.2023.3344456
Botnari A, Kadar M, Patrascu JM (2024) A comprehensive evaluation of deep learning models on knee MRIs for the diagnosis and classification of meniscal tears: a systematic review. Diagnostics 14(11):1090. https://doi.org/10.3390/diagnostics14111090
Shojaei S, Abadeh MS, Momeni Z (2023) An evolutionary explainable deep learning approach for Alzheimer’s MRI classification. Expert Syst Appl 220:119709. https://doi.org/10.1016/j.eswa.2023.119709
Benbakreti S, Benouis M, Roumane A, Benbakreti S (2023) Impact of the data augmentation on the detection of brain tumor from MRI images based on CNN and pretrained models. Multimedia Tools Appl 83(13):39459–39478. https://doi.org/10.1007/s11042-023-17092-0
Bharadwaj UU et al (2023) Deep learning for automated, interpretable classification of lumbar spinal stenosis and facet arthropathy from axial MRI. Eur Radiol 33(5):3435–3443. https://doi.org/10.1007/s00330-023-09483-6
Singh SK et al (2023) A novel deep learning-based technique for detecting prostate cancer in MRI images. Multimedia Tools Appl 83(5):14173–14187. https://doi.org/10.1007/s11042-023-15793-0
Li Q, Qiu M, Nie S, Chen F, Wang X, Zhang H (2025) Arthroscopic repair technique of lateral meniscus tear at the popliteal hiatus area. Arthroscopy Tech 14(2):102926. https://doi.org/10.1016/j.eats.2024.102926
Yang W, Li Yi, Shuyan Wu, Wei Yu, Meng C, Wang H, Huang W (2024) A Modified suture hook technique for lateral meniscus tears: Chinese union suture procedure (CUSP). Arthroscopy Tech 13(5):102946. https://doi.org/10.1016/j.eats.2024.102946
Noriega GAG, Hernández PJL, Huependo GAH, SanchezCruz DA (2024) Ramp-like lateral meniscus tear. Description of an infrequent lesion. J ISAKOS 9(4):734–739. https://doi.org/10.1016/j.jisako.2024.04.005
A. Vaswani vd., “Attention Is All You Need”, 01 Ağustos 2023, arXiv: arXiv:1706.03762. https://doi.org/10.48550/arXiv.1706.03762.
A. Dosovitskiy vd., “An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale”, 03 Haziran 2021, arXiv: arXiv:2010.11929. https://doi.org/10.48550/arXiv.2010.11929.
H. Touvron, M. Cord, M. Douze, F. Massa, A. Sablayrolles, ve H. Jégou, “Training data-efficient image transformers & distillation through attention”, 15 Ocak 2021, arXiv: arXiv:2012.12877. https://doi.org/10.48550/arXiv.2012.12877.
Ghosh S, Islam MA (2023) Performance evaluation and comparison of heart disease prediction using machine learning methods with elastic net feature selection. Am J Appl Math Stat 11(2):35–49. https://doi.org/10.12691/ajams-11-2-1
Liu J, Geng T, Jiang W, Fan S, Chen J, Jia C, Ji S (2024) A new application of Elasticnet regression based near-infrared spectroscopy model: prediction and analysis of 2,3,5,4′-tetrahydroxy stilbene-2-O-β-D-glucoside and moisture in Polygonum multiflorum. Microchem J 199:110095. https://doi.org/10.1016/j.microc.2024.110095
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc Series B: Stat Methodol 67(2):301–320. https://doi.org/10.1111/j.1467-9868.2005.00503.x
Akdemir E, Barışçı N (2024) A review on deep learning applications with semantics. Expert Syst Appl 251:124029. https://doi.org/10.1016/j.eswa.2024.124029
TanzibHosain Md, Jim JR, Mridha MF, Kabir MM (2024) Explainable AI approaches in deep learning: advancements, applications and challenges. Comput Elect Eng 117:109246. https://doi.org/10.1016/j.compeleceng.2024.109246
Patel RK, Kashyap M (2023) The study of various registration methods based on maximal stable extremal region and machine learning. Comput Methods Biomech Biomed Eng Imag Visualizat 11(6):2508–2515. https://doi.org/10.1080/21681163.2023.2243351
Wei Ze, Chen F, Liu H, Huang R, Pan K, Ji W, Wang J (2024) Mapping the application research on machine learning in the field of ionic liquids: a bibliometric analysis. Fluid Phase Equilib 583:114117. https://doi.org/10.1016/j.fluid.2024.114117
Yao C, Ren J, Bai R, Heshan Du, Liu J, Jiang X (2024) Progressively-orthogonally-mapped EfficientNet for action recognition on time-range-Doppler signature. Expert Syst Appl 255:124824. https://doi.org/10.1016/j.eswa.2024.124824
Patel RK, Kashyap M (2022) Automated screening of glaucoma stages from retinal fundus images using BPS and LBP based GLCM features. Int J Imaging Syst Technol 33(1):246–261. https://doi.org/10.1002/ima.22797
Patel RK, Kashyap M (2023) Machine learning- based lung disease diagnosis from CT images using Gabor features in Littlewood Paley empirical wavelet transform (LPEWT) and LLE. Comput Methods Biomech Biomed Eng Imag Visualizat 11(5):1762–1776. https://doi.org/10.1080/21681163.2023.2187244
Patel RK, Chouhan SS, Lamkuche HS, Pranjal P (2024) Glaucoma diagnosis from fundus images using modified Gauss-Kuzmin-distribution-based Gabor features in 2D-FAWT. Comput Electr Eng 119:109538. https://doi.org/10.1016/j.compeleceng.2024.109538
Patel RK, Kashyap M (2023) Automated diagnosis of COVID stages using texture-based Gabor features in variational mode decomposition from CT images. Int J Imaging Syst Technol 33(3):807–821. https://doi.org/10.1002/ima.22865
Nigam S, Jain R, Singh VK, Marwaha S, Arora A, Jain S (2024) EfficientNet architecture and attention mechanism-based wheat disease identification model. Procedia Comput Sci 235:383–393. https://doi.org/10.1016/j.procs.2024.04.038
F. N. Iandola, S. Han, M. W. Moskewicz, K. Ashraf, W. J. Dally, ve K. Keutzer, “SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size”, 04 Kasım 2016, arXiv: arXiv:1602.07360. https://doi.org/10.48550/arXiv.1602.07360.
Zhang J, Duan L, Luo S, Li Ke (2023) Fault diagnosis of reciprocating machinery based on improved MEEMD-SqueezeNet. Measurement 217:113026. https://doi.org/10.1016/j.measurement.2023.113026
Özdemir EY, Özyurt F (2024) Elasticnet-based vision transformers for early detection of Parkinson’s disease. Biomed Signal Process Control 101:107198. https://doi.org/10.1016/j.bspc.2024.107198
Özyurt F, Avcı E, Sert E (2020) UC-merced image classification with CNN feature reduction using wavelet entropy optimized with genetic algorithm. Traitement du Signal 37(3):347–353. https://doi.org/10.18280/ts.370301
“Niğde Ömer Halisdemir Üniversitesi Mühendislik Bilimleri Dergisi » Makale » Yerel faz niceleme ile ayak görüntülerinin kişi, yaş ve cinsiyete göre sınıflandırılması”. Erişim: 12 Mayıs 2024. [Çevrimiçi]. Erişim adresi: https://dergipark.org.tr/tr/pub/ngumuh/issue/71284/1055199
Chen, PY and Soo, VW. (2018). Humor recognition using deep learning. In Proceedings of the 2018 conference of the North American chapter of the association for computational linguistics: Human language technologies, volume 2 (short papers) (pp. 113–117).
Liu S, Wang L, Yue W (2024) An efficient medical image classification network based on multi-branch CNN, token grouping transformer and mixer MLP. Appl Soft Comput 153:111323. https://doi.org/10.1016/j.asoc.2024.111323
Rabby SF, Arafat MA, Hasan T (2024) BT-Net: An end-to-end multi-task architecture for brain tumor classification, segmentation, and localization from MRI images. Array 22:100346. https://doi.org/10.1016/j.array.2024.100346
Kılıçarslan G, Koç C, Özyurt F, Gül Y (2023) Breast lesion classification using features fusion and selection of ensemble ResNet method. Int J Imag Syst Technol 33(5):1779–1795. https://doi.org/10.1002/ima.22894
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). This work is supported by the Scientific Research Project Fund of Fırat Üniversitesi (Grant No: MF.24.24).
Author information
Authors and Affiliations
Contributions
All authors agreed on the content of the study. HG collected all the data for analysis. FO agreed on the methodology. CK and EYO completed the analysis based on agreed steps. Results and conclusions are discussed and written together. The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Ethical approval
This study was conducted in accordance with the ethical standards of the institutional research committee and the Helsinki Declaration. This study was approved by the Clinical Research Ethics Committee of Fırat University Hospital on 27.03.2024 with decision number 23359. MRI images were anonymized, free of identifying information, and used only for scientific research. In accordance with the Declaration of Helsinki, individual patient consent was not obtained in the study, but it was stated that data use was in accordance with the ethics committee within the scope of anonymized data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Genç, H., Koç, C., Yüzgeç Özdemir, E. et al. An innovative approach to classify meniscus tears by reducing vision transformers features with elasticnet approach. J Supercomput 81, 602 (2025). https://doi.org/10.1007/s11227-025-07103-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s11227-025-07103-2