Abstract
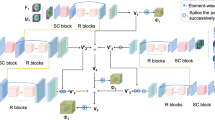
Three-dimensional (3D) medical image registration has drawn substantial research attention. In comparison to traditional approaches, deep learning techniques present significant advantages in terms of speed and accuracy. However, large deformations and complex transformations pose challenges for single-modality image registration. In this study, we propose WTDL-Net, a multi-scale registration network incorporating wavelet transform. First, low-frequency sub-images generated by WT at various resolutions are used as inputs to the multi-scale registration network. Coarse-to-fine registration is achieved by analyzing image information at different resolutions. Second, the high-frequency components derived from the WT are combined to create a high-frequency infographic. This Infographic is applied to constrain multi-level registration, thereby enhancing the optimization of registration details. The proposed approach outperforms existing deep learning-based registration techniques, as shown through comprehensive quantitative and qualitative evaluations on four MR brain scan datasets.








Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Igaki T et al (2022) Artificial intelligence-based total mesorectal excision plane navigation in laparoscopic colorectal surgery. Dis Colon Rectum. https://doi.org/10.1097/dcr.0000000000002393
Islam M, Atputharuban DA, Ramesh R, Ren H (2019) Real-time instrument segmentation in robotic surgery using auxiliary supervised deep adversarial learning. IEEE Robot Autom Lett. https://doi.org/10.1109/lra.2019.2900854
Bescos B, Facil JM, Civera J, Neira J (2018) DynaSLAM: tracking, mapping and inpainting in dynamic scenes. IEEE Robot Autom Lett. https://doi.org/10.1109/lra.2018.2860039
Li Z, Wang M (2024) Rigid point cloud registration based on correspondence cloud for image-to-patient registration in image-guided surgery. Med Phys 51(7):4554–4566. https://doi.org/10.1002/mp.17243
Wodzinski P, Banaś A, Wróbel Z, Nowak M, Borowicz M, Orzechowski M (2024) RegWSI: Whole slide image registration using combined deep feature- and intensity-based methods. Comput Methods Programs Biomed 250:108187. https://doi.org/10.1016/j.cmpb.2024.108187
Liu Y, Yao D, Zhai Z, Wang H, Chen J, Wu C, Qiao H, Li H, Shi Y (2022) Fusion of multimodality image and point cloud for spatial surface registration for knee arthroplasty. Int J Med Robot Comput Assist Surg 18(5):e2426. https://doi.org/10.1002/rcs.2426
Lecun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE. https://doi.org/10.1109/5.726791
Ren S, He K, Girshick R, Sun J (2017) Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. https://doi.org/10.1109/tpami.2016.2577031
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM. https://doi.org/10.1145/3065386
Ronneberger O, Fischer P, Brox T, U-Net: convolutional networks for biomedical image segmentation. In: lecture notes in computer science, medical image computing and computer-assisted intervention – MICCAI 2015, 2015, pp. 234–241. https://doi.org/10.1007/978-3-319-24574-4_28.
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging. https://doi.org/10.1109/tmi.2019.2897538
Jaderberg M, Simonyan K, Zisserman A, Kavukcuoglu K (2015) Spatial transformer networks. Neural information processing systems, neural information processing systems
Luo W, Li Y, Urtasun R, Zemel RS, (2016) Understanding the effective receptive field in deep convolutional neural networks. Neural Information Processing Systems, Neural Information Processing Systems
Ha IY, Wilms M, Heinrich M (2020) Semantically guided large deformation estimation with deep networks. Sensors. https://doi.org/10.3390/s20051392
ShiJ, He Y, Kong Y, Coatrieux J-L, Shu H, Yang G, and Li S (2022) "XMorpher: full transformer for deformable medical image registration via cross attention. In: medical image computing and computer-assisted intervention – MICCAI 2022, pp 217–226. https://doi.org/10.1007/978-3-031-16446-0_21.
Vaswani A et al. (2017) Attention is all you need. Neural information processing systems, neural information processing systems
Chen J, Du Y, He Y, Segars WP, Li Y, Frey E (2021) TransMorph: transformer for unsupervised medical image registration. arXiv: Image and Video Processing,arXiv: Image and Video Processing
Yuan W, Cheng J, Gong Y, He L, Zhang J (2024) MACG-net: Multi - axis cross gating network for deformable medical image registration. Comput Biol Med 178:108673. https://doi.org/10.1016/j.compbiomed.2024.108673
Zhao S, Dong Y, Chang E, Xu Y (2019) Recursive cascaded networks for unsupervised medical image registration. In: 2019 IEEE/CVF international conference on computer vision (ICCV), Seoul, Korea (South). https://doi.org/10.1109/iccv.2019.01070.
Song L, Ma M, Liu G (2023) TS-net: Two-stage deformable medical image registration network based on new smooth constraints. Magn Reson Imaging 99:26–33. https://doi.org/10.1016/j.mri.2023.01.013
Tony C, Mok W, Albert C, Chung S (2020) Large deformation diffeomorphic image registration with laplacian pyramid networks. Cornell University. arXiv, Cornell University
Wang C, Ren Q, Qin X, Yu Y (2018) Adaptive diffeomorphic multiresolution demons and their application to same modality medical image registration with large deformation. Int J Biomed Imaging 2018:1–9. https://doi.org/10.1155/2018/7314612
Chatterjee S et al (2023) Micdir: multi-scale inverse-consistent deformable image registration using UNetMSS with self-constructing graph latent. Comput Med Imaging Graph. https://doi.org/10.1016/j.compmedimag.2023.102267
Che T et al (2023) AMnet: adaptive multi-level network for deformable registration of 3D brain MR images. Med Image Anal. https://doi.org/10.1016/j.media.2023.102740
Chang Y, Li Z, Xu W (2025) CGnet: a correlation-guided registration network for unsupervised deformable image registration. IEEE Trans Med Imaging 44(3):1468–1479. https://doi.org/10.1109/TMI.2024.3505853
Zhang R et al (2025) UTSRMorph: a unified transformer and superresolution network for unsupervised medical image registration. IEEE Trans Med Imaging 44(2):891–902. https://doi.org/10.1109/TMI.2024.3467919
Li Y, Chen K, Chen C, Zhang J (2024) A bi-variant variational model for diffeomorphic image registration with relaxed Jacobian determinant constraints. Appl Math Model 130:66–93. https://doi.org/10.1016/j.apm.2024.02.033
Zhang X, Xu A, Ouyang G, Xu Z, Shen S, Chen W, Liang M, Zhang G, Wei J, Zhou X et al (2025) Wavelet-guided multi-scale ConvNeXt for unsupervised medical image registration. Bioengineering 12:406. https://doi.org/10.3390/bioengineering12040406
Li GY, Fan H (2019) Multifocus image fusion using wavelet-domain-based deep CNN. Comput Intell Neurosci 2019:1–23. https://doi.org/10.1155/2019/4179397
Lowe B, Salman H, Zhan J (2022) GHM wavelet transform for deep image super resolution
Saveljev V (2022) Continuous wavelet transform of multiview images using wavelets based on voxel patterns
Yang M, Wu F, Li W (2020) WaveletStereo: learning wavelet coefficients of disparity map in stereo matching. In: 2020 IEEE/CVF conference on computer vision and pattern recognition (CVPR), Seattle, WA, USA. https://doi.org/10.1109/cvpr42600.2020.01290.
Ghasemzadeh A, Demirel H (2018) 3D discrete wavelet transform-based feature extraction for hyperspectral face recognition. IET Biometrics. https://doi.org/10.1049/iet-bmt.2017.0082
Tian C, Zheng M, Zuo W, Zhang B, Zhang Y, Zhang D (2023) Multi-stage image denoising with the wavelet transform. Pattern Recogn. https://doi.org/10.1016/j.patcog.2022.109050
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL (2007) Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. https://doi.org/10.1162/jocn.2007.19.9.1498
Klein A et al (2009) Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. https://doi.org/10.1016/j.neuroimage.2008.12.037
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology. https://doi.org/10.2307/1932409
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. In: IEEE transactions on image processing, pp 600–612. https://doi.org/10.1109/tip.2003.819861
Thirion J-P (1998) Image matching as a diffusion process: an analogy with Maxwell’s demons. Med Image Anal. https://doi.org/10.1016/s1361-8415(98)80022-4
AVANTS B, EPSTEIN C, GROSSMAN M, GEE J (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. https://doi.org/10.1016/j.media.2007.06.004
Kim B, Kim DH, Park SH, Kim J, Lee J-G, Ye JC (2021) Cyclemorph: cycle consistent unsupervised deformable image registration. Med Image Anal. https://doi.org/10.1016/j.media.2021.102036
Cheng X, Jia X, Lu W et al (2024) WiNet: wavelet-based incremental learning for efficient medical image registration. In: proceedings of the international conference on medical image computing and computer-assisted intervention (MICCAI). Cham, Switzerland: Springer Nature Switzerland, pp. 761–771.
Funding
This work is partially funded by the Tianjin Research Project on Undergraduate Teaching Reform and Quality Construction (A231006507), the Ministry of Education's China University Industry-University-Research Innovation Fund (2022BL084), the Tianjin Municipal Education Commission Research program (2024KJ061), and the Ministry of Industry and Information Technology's Education and Examination Center's 2024 Annual Research Project.
Author information
Authors and Affiliations
Contributions
BH.C: Conceived the research idea, designed the methodology, implemented the algorithm, conducted the experiments, and analyzed the results. Drafted and revised the manuscript. BJ.Z (corresponding author): Guided the research direction, supervised the research process, optimized the algorithm design, and reviewed and revised the manuscript. B.Z: Contributed to algorithm optimization and experimental design, provided technical support, and assisted in manuscript writing and revision. CP.Z: Responsible for data preprocessing and experimental data analysis, and assisted in manuscript revision and improvement.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
This study used publicly available datasets, including OASIS, LPBA40, IBSR18, and IXI. As these datasets have already undergone ethical review and are openly accessible for research purposes, no additional ethical approval was required.
Consent to participate
This study did not involve human participants directly. The data used was obtained from publicly available sources that do not require individual consent.
Consent for publication
All authors have reviewed and approved the final manuscript for publication. Since the study is based on publicly available datasets, no additional consent for publication is required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chu, B., Zhang, B., Zhang, B. et al. WTDL-Net: medical image registration based on wavelet transform and multi-scale deep learning. J Supercomput 81, 1080 (2025). https://doi.org/10.1007/s11227-025-07567-2
Accepted:
Published:
DOI: https://doi.org/10.1007/s11227-025-07567-2