Abstract
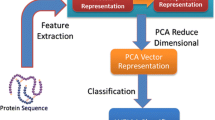
Protein–protein interaction plays an important role in biological function. Though protein interaction and non-interaction is a broad field, PPI is considered more important than PPNI. The concentration of dataset with PPNI is also used to predict the protein–protein interaction. False negatives of non-interaction data have to be identified in the non-proven negative genetic interactions. A learning approach of ensemble selection is a “build and select” strategy, where multiple classifiers have to be trained. Diversity and accuracy of the multi-classifier have to be selected to find the solution. In this paper, PPNI datasets are identified from PPI dataset. Three levels of development have been considered such as, Dataset construction carried out by Negatome, Random pair and Recombine pair methods. Feature extraction and feature selection performance can be carried out by the way of N-Gram techniques. Ensemble classification is done by utilizing the classifiers such as Support Vector Machine, Decision Tree, Neural Network and Naive Bayes. For the enhanced optimization algorithm expressed through the search operation, Genetic-PSO algorithm is proposed. The result exposes the reduced false negatives with the process of dataset construction and the execution of the random pair dataset effectively.








Similar content being viewed by others
References
Shatnawi, M. (2015). Review of Recent Protein-Protein Interaction Techniques, Emerging Trends in Computational biology, Bioinformatics and Systems Biology.
Thanos, C. D., DeLano, W. L., & Wells, J. A. (2006). Hot-spot mimicry of a cytokine receptor by a small molecule. Proceedings of the National academy of Sciences of the United States of America,103(42), 15422–15427.
Agrawal, N. J., Helk, B., & Trout, B. L. (2014). A computational tool to predict the evolutionarily conserved protein–protein interaction hot-spot residues from the structure of the unbound protein. FEBS Letters,588, 326–333.
Planas-Iglesias, J., Bonet, J., García-García, J., Marín-López, M. A., Feliu, E., & Oliva, B. (2013). Understanding protein–protein interactions using local structural features. Journal of Molecular Biology,425, 1210–1224.
Wu, S., Shao, F., Sun, R., Sui, Y., Wang, Y., & Wang, J. (2014). Analysis of human genes with protein–protein interaction network for detecting disease genes. Physica A,398, 217–228.
Peleg, O., Choi, J.-M., & Shakhnovich, E. I. (2014). Evolution of specificity in protein-protein interactions. Biophysical Journal,107, 1686–1696.
Sun, J.-T., Ao, B., Zhang, S., Bing, Z., & Yang, L. (2014). Evolving protein protein interaction networks: A model based on duplication and mutation at different rates. Journal of Theoretical Biology,350, 32–36.
Huang, Y.-A., You, Z.-H., Li, X., Chen, X., Hu, P., Li, S., et al. (2014). Construction of reliable protein–protein interaction networks using weighted sparse representation based classifier with pseudo substitution matrix representation features. Neurocomputing,218, 131–138.
Qin, H., Lu, H. H. S., Wu, W. B., & Li, W.-H. (2003). Evolution of the yeast protein interaction network. Proceedings of the National academy of Sciences of the United States of America,100(22), 12820–12824.
Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., & Sakaki, Y. (2001). Acomprehensive two-hybridanalysistoexploretheyeastproteininteractome. Proceedings of the National Academy of Sciences,98(8), 4569–4574.
Anne-Claude, G., Markus, B., Roland, K., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature,415(6868), 141–147.
Yuen, H., Albrecht, G., Adrian, H., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature,415(6868), 180–183.
De Las Rivas, J., & Fontanillo, C. (2012). Protein–protein interaction networks: Unravelling the wiring of molecular machines within the cell. Briefings in Functional Genomics:ls036.
Liu, G.-H., Shen, H.-B., & Yu, D.-J. (2016). Prediction of protein–protein interaction sites with machine-learning-based data-cleaning and post-filtering procedures. Journal of Membrane Biology,249, 141–153.
Hayat, M., & Khan, A. (2011). Predicting membrane protein types by fusing composite protein sequence features into pseudo amino acid composition. Journal of Theoretical Biology,271, 10–17.
Hayat, M., & Khan, A. (2013). WRF-TMH: Predicting transmembrane helix by fusing composition index and physicochemical properties of amino acids. AminoAcids,44, 1317–1328.
Hayat, M., & Khan, A. (2013). Prediction of membrane protein types using pseudo-aminoacid composition and ensemble classification. International Journal of Comput and Electrical Engineering,5, 456.
Ofran, Y., & Rost, B. (2003). Predicted protein-protein interaction sites from local sequence information. FEBS Letters,544, 236–239. https://doi.org/10.1016/S0014-5793(03)00456-3.
Yan, C., Dobbs, D., & Honavar, V. (2004). A two-stage classifier for identification of protein-protein interface residues. Bioinformatics,20, i371–i378. https://doi.org/10.1093/bioinformatics/bth920.
Torchala, M., Moal, I. H., Chaleil, R. A. G., Fernandez-Recio, J., & Bates, P. A. (2013). Swarm-Dock: A server for flexible protein–protein docking. Bioinformatics,29, 807–809.
Ghoorah, A. W., Devignes, M.-D., Smaïl-Tabbone, M., & Ritchie, D. W. (2011). Spatial clustering of protein binding sites for template based protein docking. Bioinformatics,27, 2820–2827.
Tuncbag, N., Gursoy, A., & Keskin, O. (2013). Identification of computational hotspots in protein interfaces: Combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics,25, 1513–1520.
Grove, L. E., Hall, D. R., Beglov, D., Vajda, S., Kozakov, D., & Flex, F. T. (2013). Accounting for binding site flexibility to improve fragment-based identification of druggable hot spots. Bioinformatics,29, 1218–1219.
Navlakha, S., & Kingsford, C. (2010). The power of protein interaction networks for associating genes with diseases. Bioinformatics,26, 1057–1063.
Mørk, S., Pletscher-Frankild, S., PallejaCaro, A., Gorodkin, J., & Jensen, L. J. (2014). Protein- driven inference of miRNA-disease associations. Bioinformatics,30, 392–397.
Zinzalla, G., & Thurston, D. E. (2009). Targeting protein–protein interactions for therapeutic intervention: A challenge for the future. Future Medicinal Chemistry,1, 65–93.
Johnson, D. K., & Karanicolas, J. (2019). Druggable protein interaction sites are more predisposed to surface pocket formation than the rest of the protein surface. PLoS Computational Biology,9(3), e1002951.
Mignani, S., ElKazzouli, S., Bousmina, M. M., & Majoral, J.-P. (2014). Den drimer space exploration: An assessment of dendrimers/dendritic scaffolding as inhibitors of protein–protein interactions, a potential new area of pharmaceutical development. Chemical Reviews,114, 1327–1342.
Huang, Y.-A., You, Z.-H., Li, X., Chen, X., Hu, P., Li, S., et al. (2016). Construction of reliable protein–protein interaction networks using weighted sparse representation based classifier with pseudo substitution matrix representation features. Neurocomputing,218, 131–138.
Shin, W.-H., Christoffer, C. W., & Kihara, D. (2017). In silico structure-based approaches to discover protein-protein interaction-targeting drugs. Methods,131, 22–32.
Zhang, J., Yang, H., Song, H., & Zhang, Y. (2017). An improved archaeology algorithm based on integrated multi-source. Biological Information for Yeast Protein Interaction Network, IEEE Access
Brun, C., Chevenet, F., Martin, D., Wojcik, J., Guénoche, A., & Jacq, B. (2003). Functional classification of proteins for the prediction of cellular function from a protein-protein interaction network. Genome Biology,5, R6.
Tahir, M., & Hayat, M. (2017). Machine learning based identification of protein–protein interactions using derived features of physiochemical properties and evolutionary profiles. Artificial Intelligence in Medicine,78, 61–71.
Jia, J., Liu, Z., Xiao, X., Liu, B., & Chou, K.-C. (2014). Identification of protein-protein binding sites by incorporating the physicochemical properties and stationary wavelet transforms into pseudo amino acid composition. Journal of Biomolecular Structure and Dynamics, ISSN: 0739-1102 (Print) 1538-0254 (Online).
Wei, Z.-S., Han, K., Yang, J.-Y., Shen, H.-B., & Yu, D.-J. (2016). Protein–protein interaction sites prediction by ensembling SVM and sample weighted random forests. Neuro Computing,193, 201–212.
Zhanga, L., Yua, G., Xiab, D., & Wang, J. (2018). Protein-protein interactions prediction based on ensemble deep neural networks. Neurocomputing,S0925–2312(18), 30633–30637.
Hu, L., & Chan, K. C. C. (2016). Extracting coevolutionary features in protein sequences for predicting protein-protein interactions. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2520923.
Qiao, Y., Xiong, Y., Gao, H., Zhu, X., & Chen, P. (2018). Protein-protein interface hot spots prediction based on a hybrid feature selection strategy. BMC Bioinformatics,19, 14.
Zahiri, J., Yaghoubi, O., Mohammad-Noori, M., Ebrahimpour, R., & Masoudi-Nejad, A. (2013). PPIevo: Protein–protein interaction prediction from PSSM based evolutionary information. Genomics,102, 237–242.
Karthik, M.N., & Davis, M. (2012). Search Using N-gram Technique Based Statistical Analysis for Knowledge Extraction in Case Based Reasoning Systems. https://arxiv.org/ftp/cs/papers/0407/0407009.pdf
Dasgupta, K., Mandal, B., Dutta, P., Mandal, J. K., & Dam, S. (2013). Agenetic algorithm GA) based load balancing strategy for cloudcomputing. Procedia Technology,10, 340–347.
Zhang, Z., & Zhang, X. (2010). A load balancing mechanism basedon ant colony and complex network theory in open cloud computing federation. In Proceedings of the 2nd International Conference on Industrial Mechatronics and Automation (ICIMA’10), vol. 2, pp. 240–243.
Braun, T. D., Siegel, H. J., Beck, N., et al. (2001). A comparison of eleven static heuristics for mapping a class of independent tasks onto heterogeneous distributed computing systems. Journal of Parallel and Distributed Computing,61(6), 810–837.
Alabdulrahman, R. (2014), A comparative study of ensemble active learning, Thesis, University of Ottawa.
Read, J., Bifet, A., Pfahringer, B., & Holmes, G. (2012). Batch-incremental versus instance-incremental learning in dynamic and evolving data. Advances in Intelligent Data Analysis XI , Springer (pp. 313–323)
Witten, I. H., & Frank, E. (2005). Data Mining: Practical machine learning Tools and techniques: Morgan Kaufmann.
Ko, A.H.-R., Sabourin, R., & de Souza Britto, Jr, A. (2007). A new dynamic ensemble selection method for numeral recognition Multiple Classifier Systems (pp. 431–439): Springer: Berlin.
Mazid, M. M., Ali, S., & Tickle, K. S. (2010). Improved C4. 5 algorithm for rule based classification. Paper presented at the proceedings of the 9th WSEAS international conference on Artificial intelligence, knowledge Engineering and data bases.
Melville, P., & Mooney, R. J. (2003). Constructing diverse classifier ensembles using artificial training examples. Paper presented at the IJCAI.
Acknowledgements
The authors would like to thank bharathiar University for providing the infrastructure to carry out the research work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lakshmi, P., Ramyachitra, D. An Improved Genetic with Particle Swarm Optimization Algorithm Based on Ensemble Classification to Predict Protein–Protein Interaction. Wireless Pers Commun 113, 1851–1870 (2020). https://doi.org/10.1007/s11277-020-07296-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11277-020-07296-0