Abstract
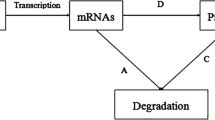
Modeling genetic regulatory networks is an important research topic in genomic research and computational systems biology. This paper considers the problem of constructing a genetic regulatory network (GRN) using the discrete dynamic system (DDS) model approach. Although considerable research has been devoted to building GRNs, many of the works did not consider the time-delay effect. Here, the authors propose a time-delay DDS model composed of linear difference equations to represent temporal interactions among significantly expressed genes. The authors also introduce interpolation scheme and re-sampling method for equalizing the non-uniformity of sampling time points. Statistical significance plays an active role in obtaining the optimal interaction matrix of GRNs. The constructed genetic network using linear multiple regression matches with the original data very well. Simulation results are given to demonstrate the effectiveness of the proposed method and model.
Similar content being viewed by others
References
S. Huang and D. Ingber, Shape-dependent control of cell growth, differentiation, and apoptosis: Switching between attractors in cell regulatory networks, Exp. Cell Res., 2000, 261: 91–103.
H. de Jong, Modeling and simulation of genetic regulatory systems: A literature review, J. Comput. Biol., 2002, 9: 69–103.
P. Smolen, D. Baxter, and J. Byrne, Mathematical modeling of gene network, Neuron, 2000, 26: 567–580.
D. Endy and R. Brent. Modeling cellular behavior, Nature, 2001, 409(6818): 391–395.
F. Christian, Lehner, H. Patrick, and O’Farrel, The roles of drosophila cyclins A and B in mitotic control, Cell, 1990, 61(3): 535–547.
J. Tyson, Modeling the cell division cycle: Cdc2 and cyclin interactions, Proc. Natl. Acad. Sci. USA, 1991, 88(16): 7328–7332.
W. Ching, E. Fung, M. Ng, and T. Akutsu, On construction of stochastic genetic networks based on gene expression sequences, International Journal of Neural Systems, 2005, 15: 297–310.
S. Zhang, W. Ching, N. Tsing, H. Leung, and D. Guo, A new multiple regression approach for the construction of genetic regulatory networks, Journal of Artificial Intelligence in Medicine, 2010, 48: 153–160.
M. Gibson and E. Mjolsness, Modeling the Activity of Single Genes, ed. by J. M. Bower, H. Bolouri, Cambridge MA: MIT Press, Chapter 1, 2001.
S. Kim, S. Imoto, and S. Miyano, Dynamic Bayesian network and nonparametric regression for nonlinear modeling of gene networks from time series gene expression data, Proc. 1st Computational Methods in Systems Biology, Lecture Note in Computer Science, 2003, 2602: 104–113.
N. Friedman, M. Linial, I. Nachman, and D. Pe’er D, Using Bayesian networks to analyze expression data, J. Comput. Biol., 2000, 7(3–4): 601–620.
S. Kauffman, Metabolic stability and epigenesis in randomly constructed gene nets, J. Theoret. Biol., 1969, 22: 437–467.
S. Kauffman, Homeostasis and differentiation in random genetic control networks, Nature, 1969, 224: 177–178.
S. Kauffman, The Origins of Order: Self-Organization and Selection in Evolution, Oxford University Press, New York, 1993.
W. Ching, S. Zhang, M. Ng, and T. Akutsu, An approximation method for solving the steady-state probability distribution of probabilistic Boolean networks, Bioinformatics, 2007, 23: 1511–1518.
I. Shmulevich, E. Dougherty, S. Kim, and W. Zhang, From Boolean to probabilistic Boolean networks as models of genetic regulatory networks, Proceedings of the IEEE, 2002, 90: 1778–1792.
I. Shmulevich and E. Dougherty, Genomic Signal Processing, Princeton University Press, Princeton, 2007.
S. Zhang, Mathematical Models and Algorithms for Genetic Regulatory Networks, The University of Hong Kong, Hong Kong, 2007.
R. Thomas and R. d’Ari, Biological Feedback, Boca Raton, FL: CRC Press, 1990.
P. Smolen, D. Baxter, and J. Byrne, Modeling transcriptional control in gene networks: Methods, recent results, and future directions, Bull. Math. Biol., 2000, 62(2): 247–292.
M. Song, Z. OuYang, and Z. Liu, Discrete dynamical system modeling for gene regulatory networks of 5-hxymethylfurfural tolerance for ethanologenic yeast, IET Syst. Biol., 2009, 3(3): 203–218.
L. J. Allen, An Introduction to Mathematical Biology, Upper Saddle River, NJ: Pearson/Prentice Hall, 2007.
J. F. Feng, J. Jost, and M. P. Qian, Networks: from Biology to Theory, Springer, London, 2007.
R. D. Bliss, R. P. Painter, and A. G. Marr, Role of feedback inhibition in stabilizing the classical operon, J. Theor. Biol., 2002, 97: 177–193.
M. S. Dasika, A. Gupta, and C. D. Maranas, A mixed integer linear programming (MILP) framework for infering time delay in gene regulatory networks, Pac. Sym. Biocomput., 2004, 9: 474–485.
R. Guthke, U. Moller, M. Hoffmann, F. Thies, and S. Topfer, Dynamic network reconstruction from gene expression data applied to immune response during bacterial infection, Bioinformatics, 2005, 21(8): 1626–1634.
D. Kincaid and W. Cheney, Numerical Analysis: Mathematics of Scientific Computing, Pacific Grove, Calif.: Brooks/Cole, 1991.
H. Jiang, W. Ching, K. Aoki-Kinoshita, and D. Guo, Delay discrete dynamical models for genetic regulatory networks, Lecture Notes in Operations Research 13, Series Editors: Ding-Zhu Du and Xiang-Sun Zhang, The Fourth International Conference on Computational Systems Biology (ISB2010), Suzhou, China, September 9–11, 2010, 13: 93–100.
Author information
Authors and Affiliations
Corresponding author
Additional information
This Research was supported in part by HKRGC Grant, HKU Strategic Theme Grant on Computational Sciences, National Natural Science Foundation of China under Grant Nos. 10971075 and 11271144.
This paper was recommended for publication by Editor Jinhu LÜ.
Rights and permissions
About this article
Cite this article
Jiang, H., Ching, WK., Aoki-Kinoshita, K.F. et al. Modeling genetic regulatory networks: a delay discrete dynamical model approach. J Syst Sci Complex 25, 1052–1067 (2012). https://doi.org/10.1007/s11424-012-0283-2
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11424-012-0283-2