Abstract
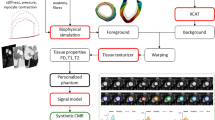
The anatomy and motion of the heart and the aorta are essential for patient-specific simulations of cardiac electrophysiology, wall mechanics and hemodynamics. Within the European integrated project euHeart, algorithms have been developed that allow to efficiently generate patient-specific anatomical models from medical images from multiple imaging modalities. These models, for instance, account for myocardial deformation, cardiac wall motion, and patient-specific tissue information like myocardial scar location. Furthermore, integration of algorithms for anatomy extraction and physiological simulations has been brought forward. Physiological simulations are linked closer to anatomical models by encoding tissue properties, like the muscle fibers, into segmentation meshes. Biophysical constraints are also utilized in combination with image analysis to assess tissue properties. Both examples show directions of how physiological simulations could provide new challenges and stimuli for image analysis research in the future.












Similar content being viewed by others
References
Aguado-Sierra J, Krishnamurthy A, Villongco C, Chuang J, Howard E, Gonzales M, Omens J, Krummen D, Narayan S, Kerckhoffs R et al (2011) Patient-specific modeling of dyssynchronous heart failure: a case study. Prog Biophys Mol Biol 107(1):147–155
Angelini E, Gerard O (2006) Review of myocardial motion estimation methods from optical flow tracking on ultrasound data. Engineering in medicine and biology society, pp 1537–1540
Avolio A, Westerhof BE, Siebes M, Tyberg JV (2009) Arterial hemodynamics and wave analysis in the frequency and time domains: an evaluation of the paradigms. Med Biol Eng Comput 47(2):107–110
Barber DC, Hose DR (2005) Automatic segmentation of medical images using image registration diagnostic and simulation applications. J Med Eng Technol 29(2):53–63
Barber DC, Oubel E, Frangi AF, Hose DR (2007) Efficient computational fluid dynamics mesh generation by image registration. Med Image Anal 11(6):648–662
Barber DC, Shi Y, Staicu C, Berrbaum P, Valverde I, Baginska J, Rutten MCM, Gaddum N, Hose DR (2011) Measurement of aortic pressure wave velocity by 4D image registration. In: Medical image understanding and analysis
Beg M, Miller M, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Bertoglio C, Moireau P, Gerbeau J (2012) Sequential parameter estimation for fluid–structure problems: application to hemodynamics. Int J Numer Methods Biomed Eng 28(4):434–455
Biesdorf A, Wörz S, Müller T, Weber TF, Heye T, Hosch W, von Tengg-Kobligk H, Rohr K (2011) Model-based segmentation and motion analysis of the thoracic aorta from 4D ECG-gated CTA images. Lecture Notes in Computer Science (MICCAI) 6891:589–596
Bousse A, Boldak C, Toumoulin C, Yang G, Laguitton S, Boulmier D (2006) Coronary extraction and characterization in multi-detector computed tomography. ITBM-RBM 27(5-6):217–226
Chabiniok R, Moireau P, Lesault PF, Rahmouni A, Deux JF, Chapelle D (2001) Trials on tissue contractility estimation from cardiac cine MRI using a biomechanical heart model. Lecture Notes in Computer Science (FIMH) 6666:304–312
Christie GR, Nielsen PM, Blackett SA, Bradley CP, Hunter PJ (2009) FieldML: concepts and implementation. Philos Trans R Soc A 367(1895):1869–1884
Daubert J, Ritter P, Le Breton H, Gras D, Leclercq CAL, Mugica J, Mabo P, Cazeau S (1998) Permanent left ventricular pacing with transvenous leads inserted into the coronary veins. PACE 21:239–245
De Craene M, Piella G, Camara O, Duchateau N, Silva E, Doltra A, D’hooge J, Brugada J, Sitges M, Frangi AF (2012) Temporal diffeomorphic free-form deformation: application to motion and strain estimation from 3d echocardiography. Med Image Anal 16(2):427–450
De Craene M, Tobón-Gomez C, Butakoff C, Duchateau N, Piella G, Rhode KS, Frangi AF (2011) Temporal diffeomorphic free form deformation (TDFFD) applied to motion and deformation quantification of tagged mri sequences. Lecture Notes in Computer Science (STACOM) 7085:68–77
Dössel O, Krueger MW, Weber FM, Wilhelms M, Seemann G (2012) Computational modeling of the human atrial anatomy and electrophysiology. Med Biol Eng Comput 50(8):773–799
Duchateau N, De Craene M, Piella G, Silva E, Doltra A, Sitges M, Bijnens B, Frangi A (2011) A spatiotemporal statistical atlas of motion for the quantification of abnormal myocardial tissue velocities. Med Image Anal 15(3):316–328
Ecabert O, Peters J, Schramm H, Lorenz C, von Berg J, Walker MJ, Vembar M, Olszewski ME, Subramanyan K, Lavi G, Weese J (2008) Automatic model-based segmentation of the heart in CT images. IEEE Trans Med Imag 27(9):1189–1201
Ecabert O, Peters J, Walker MJ, Ivanc T, Lorenz C, von Berg J, Lessick J, Vembar M, Weese J (2011) Segmentation of the heart and great vessels in CT images using a model-based adaptation framework. Med Image Anal 15(6):863–876
Eisner J (1997) State-of-the-art algorithms for minimum spanning trees: a tutorial discussion. University of Pennsylvania
Elen A, Choi H, Loeckx D, Gao H, Claus P, Suetens P, Maes F, D’hooge J (2008) Three-dimensional cardiac strain estimation using spatio-temporal elastic registration of ultrasound images: a feasibility study. IEEE Trans Med Imag 27(11):1580–1591
euHeart consortium: http://www.euheart.eu/
Ferrant M, Nabavi A, Macq B, Jolesz FA, Kikinis R, Warfield SK (2001) Registration of 3-D intraoperative MR images of the brain using a finite-element biomechanical model. IEEE Trans Med Imag 20(12):1384–1397
Groth A, Weese J, Lehmann H (2012) Robust left ventricular myocardium segmentation for multi-protocol MR. SPIE Med Imaging 8314:83142S1–83142S9
Hernández Hoyos M, Orkisz M, Roux J, Douek P (1999) Inertia-based vessel axis extraction and stenosis quantification in 3D MRA images. In: Computer assisted radiology and surgery (CARS), pp 189–193
Ho SY, Sanchez-Quintana D (2008) The importance of atrial structure and fibers. Clin Anat 22:52–63
Joldes GR, Wittek A, Warfield SK, Miller K (2012) Performing brain image warping using the deformation field predicted by a biomechanical model. Comput Biomech Med 1:89–96
Kaus MR, von Berg J, Weese J, Niessen W, Pekar V (2004) Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal 8(3):245–254
Khan A, Beg M (2008) Representation of time-varying shapes in the large deformation diffeomorphic framework. In: Biomedical imaging: from nano to macro (ISBI), pp 1521–1524
Krueger MW, Schmidt V, Tobón C, Weber FM, Lorenz C, Keller DUJ, Barschdorf H, Burdumy M, Neher P, Plank G, Rhode KS, Seemann G, Sánchez-Quintana D, Saiz J, Razavi R, Dössel O (2011) Modeling atrial fiber orientation in patient-specific geometries: a semi-automatic rule-based approach. Lecture Notes in Computer Science (FIMH) 6666:223–232
Lamata P, Niederer S, Nordsletten D, Barber DC, Roy I, Hose DR, Smith N (2011) An accurate, fast and robust method to generate patient-specific cubic Hermite meshes. Med Image Anal 15(6):801–813
Ledesma-Carbayo M, Kybic J, Desco M, Santos A, Suhling M, Hunziker P, Unser M (2005) Spatio-temporal nonrigid registration for ultrasound cardiac motion estimation.. IEEE Trans Med Imag 24(9):1113–1126
Lehmann H, Kneser R, Neizel M, Peters J, Ecabert O, Kühl H, Kelm M, Weese J (2008) Integrating viability information into a cardiac model for interventional guidance. Lecture Notes in Computer Science (FIMH) 5528:312–320
Lesage D, Angelini ED, Bloch I, Funka-Lea G (2009) A review of 3D vessel lumen segmentation techniques: models, features and extraction schemes. Med Image Anal 13(6):819–845
Lorenz C, Berg J (2006) A comprehensive shape model of the heart. Med Image Anal 10(4):657–670
Lorigo LM, Faugeras OD, Grimson WEL, Keriven R, Kikinis R, Nabavi A, Westin CF (2001) Curves: curve evolution for vessel segmentation. Med Image Anal 5:195–206
Metz C, Klein S, Schaap M, Van Walsum T, Niessen W (2011) Nonrigid registration of dynamic medical imaging data using n-d+t b-splines and a groupwise optimization approach. Med Image Anal 15(2):238–249
Nacken P, Toet A, Vincent L (1992) Graph morphology. J Vis Commun Image Represent 3(1):24–38
Neher P, Barschdorf H, Dries S, Weber FM, Krueger MW, Dössel O, Lorenz C (2011) Automatic segmentation of cardiac CTs—personalized atrial models augmented with electrophysiological structures. Lecture Notes in Computer Science (FIMH) 6666:80–87
Nickisch H, Barschdorf H, Weber FM, Krueger MW, Dössel O, Weese J (2012) From image to personalized cardiac simulation: encoding anatomical structures into a model-based segmentation framework. Lecture Notes in Computer Science (STACOM) (accepted)
Oubel E, De Craene M, Hero AO, Pourmorteza A, Huguet M, Avegliano G, Bijnens BH, Frangi AF (2012) Cardiac motion estimation by joint alignment of tagged mri sequences. Med Image Anal 16(1):339–350
Peters J, Ecabert O, Meyer C, Schramm H, Kneser R, Groth A, Weese J (2007) Automatic whole heart segmentation in static magnetic resonance image volumes. Lecture Notes in Computer Science (MICCAI) 4792:402–410
Peyrat JM, Sermesant M, Pennec X, Delingette H, Xu C, McVeigh ER, Ayache N (2007) A computational framework for the statistical analysis of cardiac diffusion tensors: application to a small database of canine hearts. IEEE Trans Med Imag 26(10):1–15
Piella G, De Craene M, Yao C, Penney GP, Frangi AF (2011) Multiview diffeomorphic registration for motion and strain estimation from 3D ultrasound sequences. Lecture Notes in Computer Science (FIMH) 6666:375–383
Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, Koller T, Gerig G, Kikinis R (1998) Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal 2(2):143–168
Sermesant M, Chabiniok R, Chinchapatnam P, Mansi T, Billet F, Moireau P, Peyrat JM, Wong K, Relan J, Rhode K, Ginks M, Lambiase P, Delingette H, Sorine M, Rinaldi CA, Chapelle D, Razavi R, Ayache N (2012) Patient-specific electromechanical models of the heart for the prediction of pacing acute effects in CRT: a preliminary clinical validation. Med Image Anal 16(1):201–215
Sermesant M, Konukoğlu E, Delingette H, Coudière Y, Chinchapatnam P, Rhode KS, Razavi R, Ayache N (2007) An anisotropic multi-front fast marching method for real-time simulation of cardiac electrophysiology. Lecture Notes in Computer Science (FIMH) 4466:160–169
Shiffman S, Rubin G, Napel S (2000) Medical image segmentation using analysis of isolable-contour maps. IEEE Trans Med Imag 19(11):1064–1074
Sigg DC, Iaizzo PA, Xiao Y-F, He B (2010) Cardiac electrophysiology methods and models. Springer, London
Smith N, de Vecchi A, McCormick M, Nordsletten D, Camara O, Frangi AF, Delingette H, Sermesant M, Relan J, Ayache N, Krueger MW, Schulze WHW, Hose R, Valverde I, Beerbaum P, Staicu C, Siebes M, Spaan J, Hunter P, Weese J, Lehmann H, Chapelle D, Razavi R (2011) euHeart: personalized and integrated cardiac care using patient-specific cardiovascular modelling. Interface Focus 1(3):349–364
Streeter D, Spontnitz H, Patel D, Ross J, Sonnenblick E (1969) Fiber orientation in the canine left ventricle during diastole and systole. Circ Res 24:339–347
Summers P, Bhalerao A, Hawkes D (1997) Multiresolution, model-based segmentation of mr angiograms. J Magn Reson Imaging 7(6):950–957
Tobón-Gomez C, De Craene M (2011) A multimodal database for the 1st cardiac motion analysis challenge. Lecture Notes in Computer Science (STACOM) 7085:33–44
Toumoulin C, Boldak C, Dillenseger JL, Coatrieux JL, Rolland Y (2001) Fast detection and characterization of vessels in very large 3-D data sets using geometrical moments. IEEE Trans Biomed Eng 48(5):604–606
Velut J, Toumoulin C, Coatrieux JL (2010) 3D coronary structure tracking algorithm with regularization and multiple hypotheses in MRI. In: Biomedical imaging: from nano to macro (ISBI), pp 37–40. Piscataway, NJ, USA
Verdonck B, Bloch L, Maitre H, Vandermeulen D, Suetens P, Marchal G (1996) Accurate segmentation of blood vessels from 3D medical images. In: International conference on image processing (ICIP) 3:311–314
Wang H, Amini A (2012) Cardiac motion and deformation recovery from MRI: a review. IEEE Trans Med Imag 31(2):487–503
Wilson DL, Noble JA (1997) Segmentation of cerebral vessels and aneurysms from mr angiography data. In: Information processing in medical imaging (IPMI), pp 423–428
Wittek A, Miller K, Kikinis R, Warfield SK (2007) Patient-specific model of brain deformation: application to medical image registration. J Biomech 40(4):919–929
Zhao F, Zhang H, Wahle A, Thomas MT, Stolpen AH, Scholz TD, Sonka M (2009) Congenital aortic disease: 4D magnetic resonance segmentation and quantitative analysis. Med Image Anal 13(3):483–493
Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D (2008) Four-chamber heart modeling and automatic segmentation for 3D cardiac CT volumes using marginal space learning and steerable features. IEEE Trans Med Imag 27(11):1668–1681
Acknowledgments
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement number 224495 (euHeart project).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weese, J., Groth, A., Nickisch, H. et al. Generating anatomical models of the heart and the aorta from medical images for personalized physiological simulations. Med Biol Eng Comput 51, 1209–1219 (2013). https://doi.org/10.1007/s11517-012-1027-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-012-1027-0