Abstract
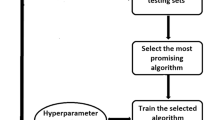
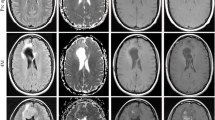
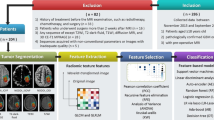
GBM is a markedly heterogeneous brain tumor consisting of three main volumetric phenotypes identifiable on magnetic resonance imaging: necrosis (vN), active tumor (vAT), and edema/invasion (vE). The goal of this study is to identify the three glioblastoma multiforme (GBM) phenotypes using a texture-based gray-level co-occurrence matrix (GLCM) approach and determine whether the texture features of phenotypes are related to patient survival. MR imaging data in 40 GBM patients were analyzed. Phenotypes vN, vAT, and vE were segmented in a preprocessing step using 3D Slicer for rigid registration by T1-weighted imaging and corresponding fluid attenuation inversion recovery images. The GBM phenotypes were segmented using 3D Slicer tools. Texture features were extracted from GLCM of GBM phenotypes. Thereafter, Kruskal–Wallis test was employed to select the significant features. Robust predictive GBM features were identified and underwent numerous classifier analyses to distinguish phenotypes. Kaplan–Meier analysis was also performed to determine the relationship, if any, between phenotype texture features and survival rate. The simulation results showed that the 22 texture features were significant with p value <0.05. GBM phenotype discrimination based on texture features showed the best accuracy, sensitivity, and specificity of 79.31, 91.67, and 98.75 %, respectively. Three texture features derived from active tumor parts: difference entropy, information measure of correlation, and inverse difference were statistically significant in the prediction of survival, with log-rank p values of 0.001, 0.001, and 0.008, respectively. Among 22 features examined, three texture features have the ability to predict overall survival for GBM patients demonstrating the utility of GLCM analyses in both the diagnosis and prognosis of this patient population.




Similar content being viewed by others
References
“3D Slicer.” http://www.slicer.org/. Accessed 20 Oct 2014
Aggarwal CC (2014) Data classification: algorithms and applications. CRC Press, Boca Raton
Al-Kadi OS, Watson D (2008) Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng 55(7):1822–1830
Bonavia R, Inda M-M, Cavenee WK, Furnari FB (2011) Heterogeneity maintenance in glioblastoma: a social network. Cancer Res 71(12):4055–4060
Brown RA, Frayne R (2008) A comparison of texture quantification techniques based on the Fourier and S transforms. Med Phys 35(11):4998–5008
Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068
Castellano G, Bonilha L, Li LM, Cendes F (2004) Texture analysis of medical images. Clin Radiol 59(12):1061–1069
Chaddad A (2015) Automated feature extraction in brain tumor by magnetic resonance imaging using gaussian mixture models. Int J Biomed Imaging 2015:e868031
Chaddad A, Tanougast C (2015) High-throughput quantification of phenotype heterogeneity using statistical features. Adv Bioinform 2015:e728164
Chaddad A, Tanougast C, Dandache A, Bouridane A (2011) Extracted haralick’s texture features and morphological parameters from segmented multispectrale texture bio-images for classification of colon cancer cells. WSEAS Trans Biol Biomed 8(2):39–50
Chaddad A, Tanougast C, Dandache A et al (2011) Improving of colon cancer cells detection based on Haralick’s features on segmented histopathological images. In: 2011 IEEE international conference on computer applications and industrial electronics (ICCAIE), pp 87–90
Chaddad A, Zinn PO, Colen RR (2014) Quantitative texture analysis for Glioblastoma phenotypes discrimination. In: 2014 International conference on control, decision and information technologies (CoDIT), pp 605–608
Chaddad A, Zinn PO, Colen RR (2014) Brain tumor identification using Gaussian mixture model features and decision trees classifier. In: 2014 48th annual conference on information sciences and systems (CISS), pp 1–4
Chen X, Wei X, Zhang Z, Yang R, Zhu Y, Jiang X (2015) Differentiation of true-progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide by GLCM texture analysis of conventional MRI. Clin Imaging 39(5):775–780
Davnall F, Yip CSP, Ljungqvist G, Selmi M, Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ, Goh V (2012) Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 3(6):573–589
Dietterich TG, Bakiri G (1995) Solving multiclass learning problems via error-correcting output codes. J Artif Int Res 2(1):263–286
Downey K, Riches SF, Morgan VA, Giles SL, Attygalle AD, Ind TE, Barton DPJ, Shepherd JH, deSouza NM (2013) Relationship between imaging biomarkers of stage I cervical cancer and poor-prognosis histologic features: quantitative histogram analysis of diffusion-weighted MR images. AJR Am J Roentgenol 200(2):314–320
Fright WR, Linney AD (1993) Registration of 3-D head surfaces using multiple landmarks. IEEE Trans Med Imaging 12(3):515–520
Fruehwald-Pallamar J, Czerny C, Holzer-Fruehwald L, Nemec SF, Mueller-Mang C, Weber M, Mayerhoefer ME (2013) Texture-based and diffusion-weighted discrimination of parotid gland lesions on MR images at 3.0 Tesla. NMR Biomed 26(11):1372–1379
Guo Y, Hastie T, Tibshirani R (2007) Regularized linear discriminant analysis and its application in microarrays. Biostatistics 8(1):86–100
Gutman DA, Cooper LAD, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD, Scarpace L, Mikkelsen T, Jain R, Wintermark M, Jilwan M, Raghavan P, Huang E, Clifford RJ, Mongkolwat P, Kleper V, Freymann J, Kirby J, Zinn PO, Moreno CS, Jaffe C, Colen R, Rubin DL, Saltz J, Flanders A, Brat DJ (2013) MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267(2):560–569
Hand DJ, Till RJ (2001) A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn 45(2):171–186
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3(6):610–621
Hearst MA, Dumais ST, Osman E, Platt J, Scholkopf B (1998) Support vector machines. IEEE Intel Syst Their Appl 13(4):18–28
Kleinbaum DG, Klein M (2012) Kaplan–Meier survival curves and the log-rank test. In: Survival analysis, 3rd edn. Springer, New York, pp 55–96
Lee J, Narang S, Martinez J, Rao G, Rao A (2015) Spatial habitat features derived from multiparametric magnetic resonance imaging data are associated with molecular subtype and 12-month survival status in glioblastoma multiforme. PLoS ONE 10(9):1–13
Lee J, Jain R, Khalil K, Griffith B, Bosca R, Rao G, Rao A (2016) Texture feature ratios from relative CBV maps of perfusion MRI are associated with patient survival in glioblastoma. Am J Neuroradiol 37(1):37–43
Levner I, Drabycz S, Roldan G, De Robles P, Cairncross JG, Mitchell R (2009) Predicting MGMT methylation status of glioblastomas from MRI texture. Medical image computing and computer-assisted intervention on MICCAI international conference of medical image computing and computer-assisted intervention, vol 12 (Pt 2), pp 522–530
Mazurowski MA, Desjardins A, Malof JM (2013) Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro Oncol 15(10):1389–1394
McCarthy BJ, Kruchko C, Dolecek TA (2013) The impact of the Benign Brain Tumor Cancer Registries Amendment Act (Public Law 107-260) on non-malignant brain and central nervous system tumor incidence trends. J Regist Manag 40(1):32–35
McKight PE, Najab J (2010) Kruskal–Wallis Test. In: The Corsini encyclopedia of psychology. Wiley, New York
Orphanidou-Vlachou E, Vlachos N, Davies NP, Arvanitis TN, Grundy RG, Peet AC (2014) Texture analysis of T1- and T2-weighted MR images and use of probabilistic neural network to discriminate posterior fossa tumours in children. NMR Biomed 27(6):632–639
Park SH, Goo JM, Jo C-H (2004) Receiver operating characteristic (ROC) Curve: practical review for radiologists. Korean J Radiol 5(1):11–18
Pope WB, Sayre J, Perlina A, Villablanca JP, Mischel PS, Cloughesy TF (2005) MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol 26(10):2466–2474
Rokach L (2007) Data mining with decision trees: theory and applications. World Scientific, Singapore
Shao J (1993) Linear model selection by Cross-validation. J Am Stat Assoc 88(422):486–494
Soh L-K, Tsatsoulis C (1999) Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans Geosci Remote Sens 37(2):780–795
Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 110(10):4009–4014
Stupp R, Hegi ME, van den Bent MJ, Mason WP, Weller M, Mirimanoff RO, Cairncross JG, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group (2006) Changing paradigms–an update on the multidisciplinary management of malignant glioma. Oncologist 11(2):165–180
Stupp R, Hegi ME, van den Bent MJ, Mason WP, Weller M, Mirimanoff RO, Cairncross JG (2006) Changing paradigms—an update on the multidisciplinary management of malignant glioma. Oncologist 11(2):165–180
Teruel JR, Heldahl MG, Goa PE, Pickles M, Lundgren S, Bathen TF, Gibbs P (2014) Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed 27(8):887–896
Turkington TG, Hoffman JM, Jaszczak RJ, MacFall JR, Harris CC, Kilts CD, Pelizzari CA, Coleman RE (1995) Accuracy of surface fit registration for PET and MR brain images using full and incomplete brain surfaces. J Comput Assist Tomogr 19(1):117–124
Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110
Wallisch P, Lusignan ME, Benayoun MD, Baker TI, Dickey AS, Hatsopoulos NG (2014) MATLAB for neuroscientists: an introduction to scientific computing in MATLAB. Academic Press, Cambridge
Zacharaki EI, Wang S, Chawla S, Soo Yoo D, Wolf R, Melhem ER, Davatzikos C (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62(6):1609–1618
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Ethical statement
The materials are in compliance with all applicable laws, regulations, and policies for the protection of medical data, and any necessary approvals, authorizations, and informed consent documents were obtained.
Rights and permissions
About this article
Cite this article
Chaddad, A., Tanougast, C. Extracted magnetic resonance texture features discriminate between phenotypes and are associated with overall survival in glioblastoma multiforme patients. Med Biol Eng Comput 54, 1707–1718 (2016). https://doi.org/10.1007/s11517-016-1461-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-016-1461-5