Abstract
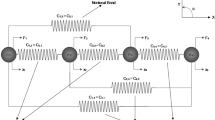
Soft tissue deformation modelling forms the basis of development of surgical simulation, surgical planning and robotic-assisted minimally invasive surgery. This paper presents a new methodology for modelling of soft tissue deformation based on reaction-diffusion mechanics via neural dynamics. The potential energy stored in soft tissues due to a mechanical load to deform tissues away from their rest state is treated as the equivalent transmembrane potential energy, and it is distributed in the tissue masses in the manner of reaction-diffusion propagation of nonlinear electrical waves. The reaction-diffusion propagation of mechanical potential energy and nonrigid mechanics of motion are combined to model soft tissue deformation and its dynamics, both of which are further formulated as the dynamics of cellular neural networks to achieve real-time computational performance. The proposed methodology is implemented with a haptic device for interactive soft tissue deformation with force feedback. Experimental results demonstrate that the proposed methodology exhibits nonlinear force-displacement relationship for nonlinear soft tissue deformation. Homogeneous, anisotropic and heterogeneous soft tissue material properties can be modelled through the inherent physical properties of mass points.

Soft tissue deformation modelling with haptic feedback via neural dynamics-based reaction-diffusion mechanics.











Similar content being viewed by others
References
Bro-Nielsen M (1998) Finite element modeling in surgery simulation. Proc IEEE 86:490–503
Zhang J, Zhong Y, Gu C (2018) Deformable models for surgical simulation: a survey. IEEE Rev Biomed Eng:1–1
Miller K (2016) Computational biomechanics for patient-specific applications. Ann Biomed Eng 44:1–2
Cover SA, Ezquerra NF, O’Brien JF, Rowe R, Gadacz T, Palm E (1993) Interactively deformable models for surgery simulation. IEEE Comput Graph Appl 13:68–75
CaniGascuel M, Desbrun M (1997) Animation of deformable models using implicit surfaces. IEEE Trans Vis Comput Graph 3:39–50
Duan Y, Huang W, Chang H, Chen W, Zhou J, Teo SK, Su Y, Chui CK, Chang S (2016) Volume preserved mass-spring model with novel constraints for soft tissue deformation. IEEE J Biomed Health Inform 20:268–280
Frisken-Gibson SF (1997) 3D ChainMail: a fast algorithm for deforming volumetric objects, Proceedings of the Symposium on Interactive 3D graphics, 149–154
Zhang J, Zhong Y, Smith J, Gu C (2016) A new ChainMail approach for real-time soft tissue simulation. Bioengineered 7:246–252
Zhang J, Zhong Y, Gu C (2017) Ellipsoid bounding region-based ChainMail algorithm for soft tissue deformation in surgical simulation. Int J Interact Des Manuf (IJIDeM)
Zhang J, Zhong Y, Smith J, Gu C (2017) ChainMail based neural dynamics modeling of soft tissue deformation for surgical simulation. Technol Health Care 25:231–239
Camara M, Mayer E, Darzi A, Pratt P (2016) Soft tissue deformation for surgical simulation: a position-based dynamics approach. Int J Comput Assist Radiol Surg 11:919–928
Misra S, Ramesh KT, Okamura AM (2008) Modeling of tool-tissue interactions for computer-based surgical simulation: a literature review. Presence Teleop Virt 17:463–491
Cotin S, Delingette H, Ayache N (1999) Real-time elastic deformations of soft tissues for surgery simulation. IEEE Trans Vis Comput Graph 5:62–73
Wu W, Heng PA (2005) An improved scheme of an interactive finite element model for 3D soft-tissue cutting and deformation. Vis Comput 21:707–716
Weber D, Mueller-Roemer J, Altenhofen C, Stork A, Fellner D (2015) Deformation simulation using cubic finite elements and efficient p-multigrid methods. Comput Graph-Uk 53 (185–195
Yang C, Li S, Lan Y, Wang L, Hao A, Qin H (2016) Coupling time-varying modal analysis and FEM for real-time cutting simulation of objects with multi-material sub-domains. Comput Aided Geom Des 43:53–67
Huang J, Liu X, Bao H, Guo B, Shum H-Y (2006) An efficient large deformation method using domain decomposition. Comput Graph-Uk 30:927–935
Strbac V, Sloten JV, Famaey N (2015) Analyzing the potential of GPGPUs for real-time explicit finite element analysis of soft tissue deformation using CUDA. Finite Elem Anal Des 105:79–89
Cotin S, Delingette H, Ayache N (2000) A hybrid elastic model for real-time cutting, deformations, and force feedback for surgery training and simulation. Vis Comput 16:437–452
Zhu B, Gu L (2012) A hybrid deformable model for real-time surgical simulation. Comput Med Imaging Graph 36:356–365
Zhang GY, Wittek A, Joldes GR, Jin X, Miller K (2014) A three-dimensional nonlinear meshfree algorithm for simulating mechanical responses of soft tissue. Eng Anal Bound Elem 42:60–66
Courtecuisse H, Allard J, Kerfriden P, Bordas SPA, Cotin S, Duriez C (2014) Real-time simulation of contact and cutting of heterogeneous soft-tissues. Med Image Anal 18:394–410
Xu S, Liu X, Zhang H, Hu L (2011) A nonlinear viscoelastic tensor-mass visual model for surgery simulation. IEEE Trans Instrum Meas 60:14–20
Dick C, Georgii J, Westermann R (2011) A real-time multigrid finite hexahedra method for elasticity simulation using CUDA. Simul Model Pract Theory 19:801–816
Miller K, Joldes G, Lance D, Wittek A (2007) Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation. Int J Numer Method Biomed Eng 23:121–134
Johnsen SF, Taylor ZA, Clarkson MJ, Hipwell J, Modat M, Eiben B, Han L, Hu Y, Mertzanidou T, Hawkes DJ, Ourselin S (2015) NiftySim: a GPU-based nonlinear finite element package for simulation of soft tissue biomechanics. Int J Comput Assist Radiol Surg 10:1077–1095
Goulette F, Chen Z-W (2015) Fast computation of soft tissue deformations in real-time simulation with hyper-elastic mass links. Comput Methods Appl Mech Eng 295:18–38
Zhong Y, Shirinzadeh B, Smith J (2008) Reaction-diffusion based deformable object simulation. Int J Image Graph 8:265–280
Sadd MH (2009) Elasticity: theory, applications, and numerics, Academic Press
Keldermann R, Nash M, Panfilov A (2009) Modeling cardiac mechano-electrical feedback using reaction-diffusion-mechanics systems. Physica D 238:1000–1007
Keldermann RH, Nash MP, Gelderblom H, Wang VY, Panfilov AV (2010) Electromechanical wavebreak in a model of the human left ventricle. Am J Phys Heart Circ Phys 299:H134–H143
Gizzi A, Cherubini C, Filippi S, Pandolfi A (2015) Theoretical and numerical modeling of nonlinear electromechanics with applications to biological active media. Commun Comput Phys 17:93–126
Murray JD (2002) Mathematical biology I: an introduction, vol. 17 of Interdisciplinary Applied Mathematics. Springer, New York
Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117:500–544
Luo C-h, Rudy Y (1994) A dynamic model of the cardiac ventricular action potential. I. Simulations of ionic currents and concentration changes. Circ Res 74:1071–1096
Noble D (1962) A modification of the Hodgkin–Huxley equations applicable to Purkinje fibre action and pacemaker potentials. J Physiol 160:317–352
Chua LO, Roska T (1993) The CNN paradigm. IEEE Trans Circuits Syst I, Fundam Theory Appl 40:147–156
Thiran P, Setti G, Hasler M (1998) An approach to information propagation in 1-D cellular neural networks—part I: local diffusion. IEEE Trans Circuits Syst I, Fundam Theory Appl 45:777–789
Setti G, Thiran P, Serpico C (1998) An approach to information propagation in 1-D cellular neural networks—part II: global propagation. IEEE Trans Circuits Syst I, Fundam Theory Appl 45:790–811
Kozek T, Chua LO, Roska T, Wolf D, Tetzlaff R, Puffer F, Lotz K (1995) Simulating nonlinear waves and partial differential equations via CNN—part II: typical examples. IEEE Trans Circuits Syst I, Fundam Theory Appl 42:816–820
Szolgay P, Vörös G, Erőss G (1993) On the applications of the cellular neural network paradigm in mechanical vibrating systems. IEEE Trans Circuits Syst I, Fundam Theory Appl 40:222–227
Chua LO, Yang L (1988) Cellular neural networks: theory. IEEE Trans Circuits Syst 35:1257–1272
Vijayan P, Kallinderis Y (1994) A 3D finite-volume scheme for the Euler equations on adaptive tetrahedral grids. J Comput Phys 113:249–267
Chua LO, Hasler M, Moschytz GS, Neirynck J (1995) Autonomous cellular neural networks: a unified paradigm for pattern formation and active wave propagation. IEEE Trans Circuits Syst I, Fundam Theory Appl 42:559–577
Fung Y-C (1993) Biomechanics: mechanical properties of living tissues, Springer-Verlag
Taylor ZA, Cheng M, Ourselin S (2008) High-speed nonlinear finite element analysis for surgical simulation using graphics processing units. IEEE Trans Med Imaging 27:650–663
Sparks JL, Vavalle NA, Kasting KE, Long B, Tanaka ML, Sanger PA, Schnell K, Conner-Kerr TA (2015) Use of silicone materials to simulate tissue biomechanics as related to deep tissue injury. Adv Skin Wound Care 28:59–68
Jingya Z, Jiajun W, Xiuying W, Dagan F (2014) The adaptive FEM elastic model for medical image registration. Phys Med Biol 59:97–118
Misra J, Saha I (2010) Artificial neural networks in hardware a survey of two decades of progress. Neurocomputing 74:239–255
Ullah Z, Augarde CE (2013) Finite deformation elasto-plastic modelling using an adaptive meshless method. Comput Struct 118:39–52
Picinbono G, Lombardo JC, Delingette H, Ayache N (2002) Improving realism of a surgery simulator: linear anisotropic elasticity, complex interactions and force extrapolation. J Vis Comput Animat 13:147–167
Xia P (2016) New advances for haptic rendering: state of the art. Vis Comput:1–17
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J., Zhong, Y. & Gu, C. Soft tissue deformation modelling through neural dynamics-based reaction-diffusion mechanics. Med Biol Eng Comput 56, 2163–2176 (2018). https://doi.org/10.1007/s11517-018-1849-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-018-1849-5