Abstract
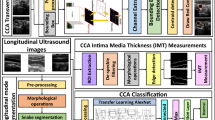
Manual ultrasound (US)-based methods are adapted for lumen diameter (LD) measurement to estimate the risk of stroke but they are tedious, error prone, and subjective causing variability. We propose an automated deep learning (DL)-based system for lumen detection. The system consists of a combination of two DL systems: encoder and decoder for lumen segmentation. The encoder employs a 13-layer convolution neural network model (CNN) for rich feature extraction. The decoder employs three up-sample layers of fully convolution network (FCN) for lumen segmentation. Three sets of manual tracings were used during the training paradigm leading to the design of three DL systems. Cross-validation protocol was implemented for all three DL systems. Using the polyline distance metric, the precision of merit for three DL systems over 407 US scans was 99.61%, 97.75%, and 99.89%, respectively. The Jaccard index and Dice similarity of DL lumen segmented region against three ground truth (GT) regions were 0.94, 0.94, and 0.93 and 0.97, 0.97, and 0.97, respectively. The corresponding AUC for three DL systems was 0.95, 0.91, and 0.93. The experimental results demonstrated superior performance of proposed deep learning system over conventional methods in literature.

ᅟ













Similar content being viewed by others
References
Ward H et al (2012) Oxford handbook of epidemiology for clinicians. OUP Oxford
Sobieszczyk P, Beckman J (2006) Carotid artery disease. Circulation 114(7):e244–e247
“What is a stroke?”. www.nhlbi.nih.gov/health/health-topics/topics/stroke. June 22, (2016)
Bots ML, Grobbee DE, Hofman A, Witteman JCM (2005) Common carotid intima-media thickness and risk of acute myocardial infarction: the role of lumen diameter. Stroke 36(4):762–767
Bots ML, Hoes AW, Koudstaal PJ, Hofman A, Grobbee DE (1997) Common carotid intima-media thickness and risk of stroke and myocardial infarction: the Rotterdam Study. Circulation 96(5):1432–1437
Tell GS, Polak JF, Ward BJ, Kittner SJ, Savage PJ, Robbins J (1994) Relation of smoking with carotid artery wall thickness and stenosis in older adults. The Cardiovascular Health Study. The Cardiovascular Health Study (CHS) Collaborative Research Group. Circulation 90(6):2905–2908
Polak JF, O'Leary DH, Kronmal RA, Wolfson SK, Bond MG, Tracy RP, Gardin JM, Kittner SJ, Price TR, Savage PJ (1993) Sonographic evaluation of carotid artery atherosclerosis in the elderly: relationship of disease severity to stroke and transient ischemic attack. Radiology 188(2):363–370
Nicolaides AN, Kakkos SK, Kyriacou E, Griffin M, Sabetai M, Thomas DJ, Tegos T, Geroulakos G, Labropoulos N, Doré CJ, Morris TP, Naylor R, Abbott AL, Asymptomatic Carotid Stenosis and Risk of Stroke (ACSRS) Study Group (2010) Asymptomatic internal carotid artery stenosis and cerebrovascular risk stratification. J Vasc Surg 52(6):1486–1496
Mann JM, Davies MJ (1996) Vulnerable plaque: relation of characteristics to degree of stenosis in human coronary arteries. Circulation 94(5):928–931
Dodge JT, Greg Brown B, Bolson EL, Dodge HT (1992) Lumen diameter of normal human coronary arteries. Influence of age, sex, anatomic variation, and left ventricular hypertrophy or dilation. Circulation 86(1):232–246
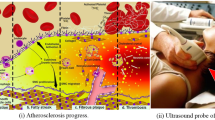
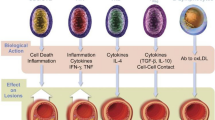
Sakakura K, Nakano M, Otsuka F, Ladich E, Kolodgie FD, Virmani R (2013) Pathophysiology of atherosclerosis plaque progression. Heart, Lung and Circulation 22(6):399–411
Sarkar S, Ghosh S, Ghosh SK, Collier A (2007) Role of transcranial Doppler ultrasonography in stroke. Postgrad Med J 83(985):683–689
Branas CC, Weingarten MS, Czeredarczuk M, Schafer PF (1994) Examination of carotid arteries with quantitative color Doppler flow imaging. J Ultrasound Med 13(2):121–127
Mitchell DG (1990) Color Doppler imaging: principles, limitations, and artifacts. Radiology 177(1):1–10
Mehra S (2010) Role of duplex Doppler sonography in arterial stenoses. Journal Indian Academy of Clinical Medicine 11(4):294–299
Jones SA, Leclerc H, Chatzimavroudis GP, Kim YH, Scott NA, Yoganathan AP (1996) The influence of acoustic impedance mismatch on poststenotic pulsed-doppler ultrasound measurements in a coronary artery model. Ultrasound Med Biol 22(5):623–634
Hoeks APG, Brands PJ, Willigers JM, Reneman RS (1999) Non-invasive measurement of mechanical properties of arteries in health and disease. Proc Inst Mech Eng H J Eng Med 213(3):195–202
Wendelhag I, Gustavsson T, Suurküla M, Berglund G, Wikstrand J (1991) Ultrasound measurement of wall thickness in the carotid artery: fundamental principles and description of a computerized analysing system. Clin Physiol Funct Imaging 11(6):565–577
Molinari F, Zeng G, Suri JS (2010) A state of the art review on intima–media thickness (IMT) measurement and wall segmentation techniques for carotid ultrasound. Comput Methods Prog Biomed 100(3):201–221
Golemati S, Stoitsis J, Sifakis EG, Balkizas T, Nikita KS (2007) Using the Hough transform to segment ultrasound images of longitudinal and transverse sections of the carotid artery. Ultrasound Med Biol 33(12):1918–1932
Molinari F, Zeng G, Suri JS (2010) An integrated approach to computer based automated tracing and its validation for 200 common carotid arterial wall ultrasound images. J Ultrasound Med 29(3):399–418
Loizou CP, Marios P (2013) Integrated system for the complete segmentation of the common carotid artery bifurcation in ultrasound images. Artificial Intelligence Applications and Innovations 412(1):292–301
Suri JS, Liu K, Singh S et al (2002) Shape recovery algorithms using level sets in 2-D/3-D medical imagery: a state-of-the-art review. IEEE Trans Inf Technol Biomed 6(1):8–28
Suri JS, Laxminarayan S (2002) PDE and level sets. Springer Science & Business Media
Araki T, Kumar PK, Suri HS, Ikeda N, Gupta A, Saba L et al (2016) Two automated techniques for carotid lumen diameter measurement: regional versus boundary approaches. J Med Syst 40(7):1–19
Kumar PK, Araki T, Rajan J, Saba L, Lavra F, Ikeda N, Sharma AM et al (2017) Accurate lumen diameter measurement in curved vessels in carotid ultrasound: an iterative scale-space and spatial transformation approach. Med Biol Eng Comput:1–20
Kuppili V et al (2017) Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J Med Syst 41(10):152
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. Proc IEEE Conf Comput Vis Pattern Recognit
Teichmann M et al (2016) MultiNet: real-time joint semantic reasoning for autonomous driving. arXiv preprint arXiv:1612.07695
Molinari F, Meiburger KM, Saba L, Zeng G, Acharya UR, Ledda M, Nicolaides A, Suri JS (2012) Fully automated dual-snake formulation for carotid intima-media thickness measurement. J Ultrasound Med 31(7):1123–1136
Iglesias JE, Sabuncu MR (2015) Multi-atlas segmentation of biomedical images: a survey. Med Image Anal 24(1):205–219
Ciresan D et al (2012) Deep neural networks segment neuronal membranes in electron microscopy images. Advances in neural information processing systems
Bar Y et al (2015) Chest pathology detection using deep learning with non-medical training. Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on. IEEE
Simonyan K and Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Hatipoglu N, Bilgin G (2017) Cell segmentation in histopathological images with deep learning algorithms by utilizing spatial relationships. Med Biol Eng Comput 55(10):1829–1848
Zhao J, Zhang M, Zhou Z, Chu J, Cao F (2017) Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Comput 55(8):1287–1301
García-Zapirain B, Elmogy M, El-Baz A, and Elmaghraby AS (2017) Classification of pressure ulcer tissues with 3D convolutional neural network. Med Biol Eng Comput 1–14
Suri JS, Haralick RM, Sheehan FH (2000) Greedy algorithm for error correction in automatically produced boundaries from low contrast ventriculograms. Pattern Anal Appl 3(1):39–60
Molinari F, Krishnamurthi G, Rajendra Acharya U, Vinitha Sree S, Zeng G, Saba L, Nicolaides A, Suri JS (2012) Hypothesis validation of far-wall brightness in carotid-artery ultrasound for feature-based IMT measurement using a combination of level-set segmentation and registration. IEEE Trans Instrum Meas 61(4):1054–1063
Gutierrez MA, Pilon PE, Lage SG, Kopel L, Carvalho RT and Furuie SS (2002) Automatic measurement of carotid diameter and wall thickness in ultrasound images. Comput Cardiol 359–362
Sahani AK, Joseph J and Sivaprakasam M (2013) Automatic measurement of lumen diameter of carotid artery in A-mode ultrasound. In Engineering in Medicine and Biology Society (EMBC), 2013 35th Annual International Conference of the IEEE, pp 3873–3876
Saba L, Araki T, Krishna Kumar P, Rajan J, Lavra F, Ikeda N, Sharma AM, Shafique S, Nicolaides A, Laird JR, Gupta A (2016) Carotid inter-adventitial diameter is more strongly related to plaque score than lumen diameter: an automated tool for stroke analysis. J Clin Ultrasound 44(4):210–220
Araki T, Kumar AM, Krishna Kumar P, Gupta A, Saba L, Rajan J, Lavra F, Sharma AM, Shafique S, Nicolaides A, Laird JR (2016) Ultrasound-based automated carotid lumen diameter/stenosis measurement and its validation system. Journal for Vascular Ultrasound 40(3):120–134
Acknowledgments
The authors at the National Institute of Technology, Goa, India, would like to acknowledge MediaLab Asia, Ministry of Electronics and Information Technology, and the Government of India for their kind support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The ethics approval was granted by Toho University IRB, Japan. Informed consent was obtained from all the patients.
Appendices
Appendix 1 Statistical test results
Appendix 2 Polyline distance method
1.1 Polyline distance metric
Polyline distance metric (PDM) [40] is used to measure the lumen diameter (LD), LI-far error, and LI-near error. In here, we focus on deriving the PDM given two border contours. Let the first and second contours be denoted as I1 and I2. Let the reference point on I1 be vertex A1 and the segment in I2 be defined by vertices A2 and A3. Let the distance between A1 and A2 be d1and the distance between A1 and A3 be denoted as d2. Let D(A1, L) be the polyline distance between vertex A1 : (x1, y1) on I1 and line segment L formed by two points A2 : (x2, y2)and A3 : (x3, y3). Let phi (φ) be the distance of the reference point, A1 towards the line segment L. The perpendicular distance between the line segment L and the reference point, A1, is given by dP. Then, the polyline distance D(A1, L) can be defined as:
where,
and
The process to obtain D(A1, L) is repeated for the rest of the points of the contour Ij and is given by:
where, N is the total number of points on I1 and \( {S}_{I_2} \) is the segment on contour I2. This algorithm is repeated in reverse, where I2 becomes the reference contour and I1 becomes the segment contour. The reverse is represented as D(I2, I1). Finally, by combining both D(I1, I2) and D(I2, I1), we obtain the PDM which is given by:
Appendix 3 Figure-of-merit and precision-of-merit
1.1 LD and mean LD computation
The LD error is computed as PDM between the ground truth LD (LDgt) and deep learning LD (LDdl). The LD for a patient i is computed as a PDM between LI-far ( LIfar(i)) and LI-near ( LInear(i)) wall of the patient. The ground truth LD (LDgt(i)) for patient i is given as:
Similarly, deep learning LD (LDdl(i)) for image i is given as:
The mean LD can therefore be computed as:
1.2 LD error and mean LD error
The LD error (εLD(i)) for an image i is computed as the absolute difference between LDgt(i) and LDdl(i) and is mathematically represented as:
If εLD(i) represents the LD error for an image i, then the mean LD error (\( {\overline{\varepsilon}}_{\mathrm{LD}} \)) for all N patients is given by:
1.3 Precision-of-merit
Using Eqs. (C.1) and (C.2), one can therefore define mathematically the precision-of-merit (POM) and is given as:
1.4 Figure-of-merit
The central tendency of the LD distribution can also be used to tell the difference between the DL-based LD and GT-based LD. Using (C.3) and (C.4), one can therefore compute figure-of-merit (FoM) and can be expressed mathematically as:
Appendix 4 LI-far and LI-near position errors
1.1 LI-far error
The LI-far error (εfar(i)) for patient i is computed as the PDM between the GT LI-far wall (\( {\mathrm{LI}}_{\mathrm{far}(i)}^{\mathrm{gt}} \)) and DL LI-far (\( {\mathrm{LI}}_{\mathrm{far}(i)}^{\mathrm{dl}} \)) wall for the patient, which is given by:
If εfar(i) represents the LI-far error for the patient i, then, the mean LI-far error (\( {\overline{\varepsilon}}_{\mathrm{far}} \)) for all N patients is given by:
1.2 LI-near error
Similarly, the LI-near error (εnear(i)) is computed as the PDM between the GT LI-near wall (\( {\mathrm{LI}}_{\mathrm{near}(i)}^{\mathrm{gt}} \)) and DL LI-near (\( {\mathrm{LI}}_{\mathrm{near}(i)}^{\mathrm{dl}} \)) wall for patient i is given by:
The mean LI-near error (\( {\overline{\varepsilon}}_{\mathrm{near}} \)) for all N patients is given by:
The corresponding symbol table is given in Appendix 5, Table 8.
Appendix 5 Symbol table
Rights and permissions
About this article
Cite this article
Biswas, M., Kuppili, V., Saba, L. et al. Deep learning fully convolution network for lumen characterization in diabetic patients using carotid ultrasound: a tool for stroke risk. Med Biol Eng Comput 57, 543–564 (2019). https://doi.org/10.1007/s11517-018-1897-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-018-1897-x